HOME
>
試薬
>
細胞情報伝達
>
エピジェネティクス
>
DNAメチル化
>
MethylFlash Methylated DNA (5-mC) Quantification Kit
HOME
>
試薬
>
遺伝子工学
>
エピジェネティクス
>
DNAメチル化
>
MethylFlash Methylated DNA (5-mC) Quantification Kit
DNA試料中の5-メチル化シトシン(5-mC)定量キット MethylFlash Methylated DNA (5-mC) Quantification Kit
掲載日情報:2016/06/14 現在Webページ番号:4728
DNA試料中のメチル化シトシン(5-mC)の総量を定量できるキットです。プレートにDNA 試料を捕捉後、メチル化DNA特異的抗体と発色または蛍光基質を用いて検出します。
※改良品のMethylFlash Easy Kitも併せてご利用下さい。従来品と比較し、より速く、より安定で、より正確な測定が可能です。
⇒改良品のMethylFlash Easy Kitはこちら。
追加しました。
特長
- 非修飾シトシンと交差せず、また 5-ハイドロメチル化シトシン(5-hmC)との交差はほとんどありません。そのため、MethylFlash Hydroxymethylated DNA Quantification Kit と組み合わせて用いることにより、試料中の 5-mCおよび5-ヒドロキシメチル化シトシン(5-hmC) の総量を定量できます。
- プレートの乾燥およびブロッキング操作が不要で、従来の製品より操作が簡便になっています。
- バックグラウンドが低く、一貫性のある結果が得られます。
- わずか 0.2 ng(Colorimetric Kit)または 50 pg(Fluorometric Kit)のメチル化DNAを定量できます。
- 分かりやすい操作法で、約 4 時間で測定できます。
- 哺乳動物、植物、菌類、バクテリア、ウイルスなど幅広い生物種由来のメチル化DNAを測定できます。
| 商品コード | P-1034 | P-1035 |
|---|---|---|
| 測定法 | 比色 | 蛍光 |
| 必要試料量 | 50~200 ng | 20~200 ng |
| 標準曲線範囲 | 0.5~10 ng | 0.2~10 |
| 測定波長 | 450 nm | 励起530 nm/蛍光590 nm |
追加しました。
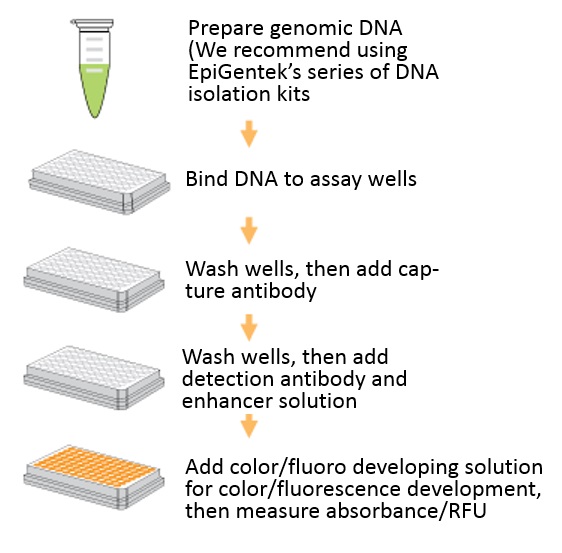
操作方法概略
追加しました。
使用例
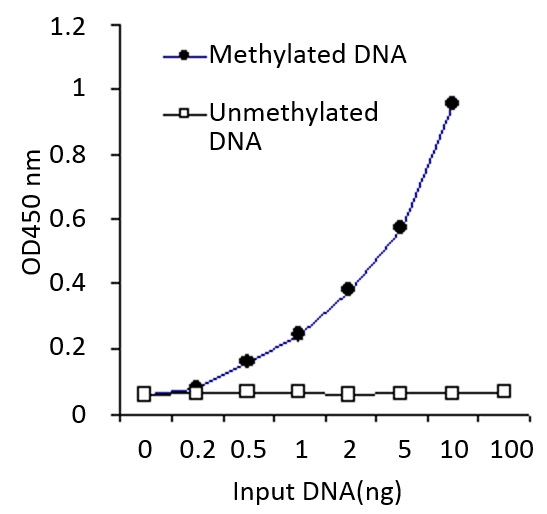
Demonstration of high sensitivity and specificity of methylated DNA (5mC) detection achieved by the MethylFlash kit. Synthetic unmethylated DNA (contains 50% of cytosine) and methylated DNA (contains 50% of 5-methylcytosine) were added into the assay wells at different concentrations and then measured with the MethylFlash™ Methylated DNA Quantification Kit (Colorimetric).
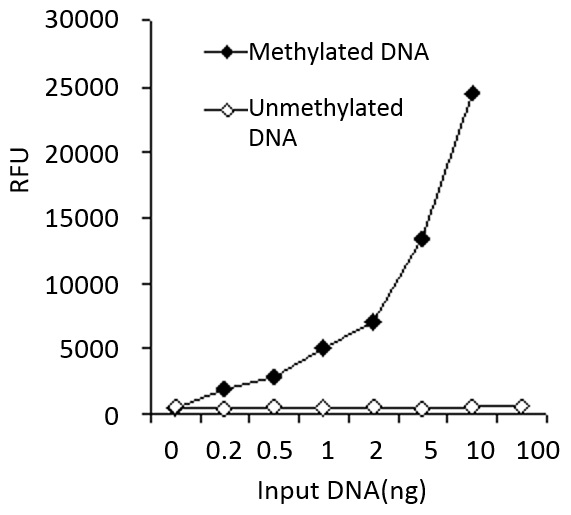
Demonstration of high sensitivity and specificity of methylated DNA detection achieved by the MethylFlash kit. Synthetic unmethylated DNA (contains 50% of cytosine) and methylated DNA (contains 50% of 5-methylcytosine) were added into the assay wells at different concentrations and then measured with the MethylFlash™ Methylated DNA Quantification Kit (Fluorometric).
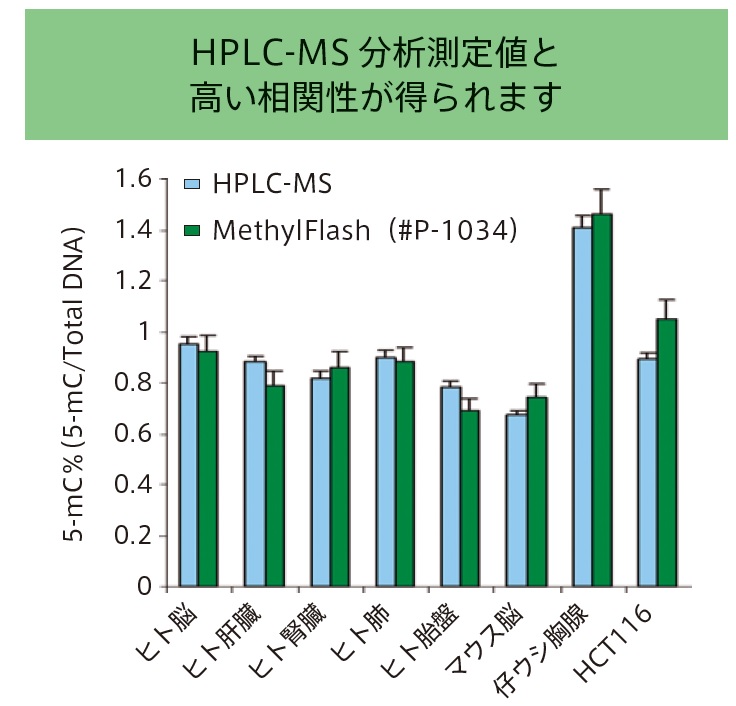
5-mC content levels in various sample types measured by MethylFlash technology versus HPLC-MS/MS, demonstrating a reliable correlation between the two techniques.
追加しました。
使用例動画
追加しました。
Epigentek Group社のDNAメチル化解析キットシリーズ
ELISAに類似した手法で、5-メチル化シトシン(5-mC)をはじめとした各種修飾核酸を定量できるキットのシリーズです。製品名をクリックすると詳細をご覧になれます。
| 測定物 | 測定試料 | 検出法 | 特長 | 製品名 |
|---|---|---|---|---|
| 5-メチル化シトシン (5-mC) |
抽出したDNA | 比色 | より迅速・簡便・高感度に検出 | MethylFlash Global DNA Methylation (5-mC) ELISA Easy Kit |
| 比色 または 蛍光 | 使用文献多数あり | MethylFlash Methylated DNA (5-mC) Quantification Kit | ||
| 5-ヒドロキシメチル化シトシン (5-hmC) |
抽出したDNA | 比色 | より迅速・簡便・高感度に検出 | MethylFlash Global DNA Hydroxymethylation (5-hmC) ELISA Easy Kit |
| 比色 または 蛍光 | 使用文献多数あり | MethylFlash Hydroxymethylated DNA (5-hmC) Quantification Kit | ||
| 5-ホルミル化シトシン (5-fC) |
抽出したDNA | 比色 | シトシン、5-mC、5-hmC、5-caCとの交差なし | MethylFlash 5-Formylcytosine (5-fC) DNA Quantification Kit |
| 5-methyl-2-deoxycytidineと 5-methylcytidineの総量 |
ヒトまたは動物の尿 | 比色 または 蛍光 | メチル化DNAおよびメチル化RNAの分解量の総量を反映 | MethylFlash Urine 5-Methylcytosine (5-mC) Quantification Kit |
追加しました。
キット内容
■共通
- Wash buffer
- Binding solution
- Negative control
- Positive control
- Capture antibody
- Detection antibody
- 8-Well assay strips with frame
- Enhancer solution
■Colorimetric Kit のみ
- Developer solution
- Stop solution
■Fluorometric Kit のみ
- Fluoro developer
- Fluoro dilutor
- Fluoro enhancer
追加しました。
価格
[在庫・価格 :2026年02月23日 00時00分現在]
| 詳細 | 商品名 |
|
文献数 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Methylated DNA Quantification Kit, Colorimetric, MethylFlash (48assays) |
|
419 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Methylated DNA Quantification Kit, Colorimetric, MethylFlash (96assays) |
|
419 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Methylated DNA Quantification Kit, Fluorometric, MethylFlash (48assays) |
|
38 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Methylated DNA Quantification Kit, Fluorometric, MethylFlash (96assays) |
|
38 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
[在庫・価格 :2026年02月23日 00時00分現在]
Methylated DNA Quantification Kit, Colorimetric, MethylFlash (48assays)
文献数: 419
- 商品コード:P-1034-48
- メーカー:EPG
- 包装:1kit
- 価格:¥92,000
- 在庫:無(未発注)
- 納期:10日程度 ※※ 表示されている納期は弊社に在庫がなく、取り寄せた場合の目安納期となります。
- 法規制等:
| 説明文 | DNA試料中のメチル化シトシンの総量を定量できるキット(5-mC Kit)。測定試料:DNA,測定範囲:0.5-10.0 ng/μl,感度:0.2 ng,検出方法:呈色,測定波長:450 nm,アッセイ法:Quantification,アッセイ数:48 assays |
||||||
|---|---|---|---|---|---|---|---|
| 別包装品 | 別包装品あり | ||||||
| 法規制等 | |||||||
| 保存条件 | 4℃,-20℃,暗所保存 | 法規備考 | |||||
| 掲載カタログ |
ニュース2021年3月15日号 p.5
|
||||||
| 製品記事 | |||||||
| 関連記事 | |||||||
Methylated DNA Quantification Kit, Colorimetric, MethylFlash (96assays)
文献数: 419
- 商品コード:P-1034-96
- メーカー:EPG
- 包装:1kit
- 価格:¥170,000
- 在庫:無(未発注)
- 納期:10日程度 ※※ 表示されている納期は弊社に在庫がなく、取り寄せた場合の目安納期となります。
- 法規制等:
| 説明文 | DNA試料中のメチル化シトシンの総量を定量できるキット(5-mC Kit)。測定試料:DNA,測定範囲:0.5-10.0 ng/μl,感度:0.2 ng,検出方法:呈色,測定波長:450 nm,アッセイ法:Quantification,アッセイ数:96 assays |
||||||
|---|---|---|---|---|---|---|---|
| 別包装品 | 別包装品あり | ||||||
| 法規制等 | |||||||
| 保存条件 | 4℃,-20℃,暗所保存 | 法規備考 | |||||
| 掲載カタログ |
ニュース2022年11月1日号 p.19 ニュース2021年3月15日号 p.5
|
||||||
| 製品記事 | |||||||
| 関連記事 | |||||||
Methylated DNA Quantification Kit, Fluorometric, MethylFlash (48assays)
文献数: 38
- 商品コード:P-1035-48
- メーカー:EPG
- 包装:1kit
- 価格:¥101,000
- 在庫:無(未発注)
- 納期:10日程度 ※※ 表示されている納期は弊社に在庫がなく、取り寄せた場合の目安納期となります。
- 法規制等:
| 説明文 | DNA試料中のメチル化シトシンの総量を定量できるキット(5-mC Kit)。測定試料:DNA,感度:50 pg,測定範囲:0.5-10.0 ng/μl,検出方法:蛍光,測定波長:530/580 nm,アッセイ法:Quantification,アッセイ数:48 assays |
||||||
|---|---|---|---|---|---|---|---|
| 別包装品 | 別包装品あり | ||||||
| 法規制等 | |||||||
| 保存条件 | 4℃,-20℃,暗所保存 | 法規備考 | |||||
| 掲載カタログ |
ニュース2021年3月15日号 p.5
|
||||||
| 製品記事 | |||||||
| 関連記事 | |||||||
Methylated DNA Quantification Kit, Fluorometric, MethylFlash (96assays)
文献数: 38
- 商品コード:P-1035-96
- メーカー:EPG
- 包装:1kit
- 価格:¥177,000
- 在庫:無(未発注)
- 納期:10日程度 ※※ 表示されている納期は弊社に在庫がなく、取り寄せた場合の目安納期となります。
- 法規制等:
| 説明文 | DNA試料中のメチル化シトシンの総量を定量できるキット(5-mC Kit)。測定試料:DNA,感度:50 pg,測定範囲:0.5-10.0 ng/μl,検出方法:蛍光,測定波長:530/580 nm,アッセイ法:Quantification,アッセイ数:96 assays |
||||||
|---|---|---|---|---|---|---|---|
| 別包装品 | 別包装品あり | ||||||
| 法規制等 | |||||||
| 保存条件 | 4℃,-20℃,暗所保存 | 法規備考 | |||||
| 掲載カタログ |
ニュース2021年3月15日号 p.5 ニュース2022年11月1日号 p.19
|
||||||
| 製品記事 | |||||||
| 関連記事 | |||||||
追加しました。
関連製品 TET1 Protein (Active)
N末端にFLAGタグが付加された組換え体TET1 (Ten-eleven translocation methylcytosine dioxygenase 1)タンパク質です。
[在庫・価格 :2026年02月23日 00時00分現在]
| 詳細 | 商品名 |
|
文献数 | ||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
TET1 Protein (Active) |
|
3 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||
[在庫・価格 :2026年02月23日 00時00分現在]
TET1 Protein (Active)
文献数: 3
- 商品コード:E12002-1
- メーカー:EPG
- 包装:10μg
- 価格:¥55,000
- 在庫:無(未発注)
- 納期:10日程度 ※※ 表示されている納期は弊社に在庫がなく、取り寄せた場合の目安納期となります。
- 法規制等:カルタヘナ該当品
| 説明文 | N末端にFLAGタグが付加された組換え体TET1 (Ten-eleven translocation methylcytosine dioxygenase 1)タンパク質。酵素カイネティックス解析,阻害物質のスクリーニング,選択的プロファイリングなどに有用。産生:Baculovirus,由来:Human |
||
|---|---|---|---|
| 法規制等 | カルタヘナ該当品 | ||
| 保存条件 | -80℃ | 法規備考 | |
| 掲載カタログ |
|
||
| 製品記事 | TET1 Protein (Active) |
||
| 関連記事 | |||
追加しました。
使用文献
| 2016年 |
|---|
| Wei H et. al. (2016). Redox/methylation mediated abnormal DNA methylation as regulators of ambient fine particulate matter-induced neurodevelopment related impairment in human neuronal cells. Sci Rep. 6:33402. |
| Ann Manzardo et. al. (2016). Examination of Global Methylation and Targeted Imprinted Genes in Prader- Willi Syndrome Imedpub. 2(3) |
| Subbanna S et. al. (2016). A single day of 5-azacytidine exposure during development induces neurodegeneration in neonatal mice and neurobehavioral deficits in adult mice. Physiol Behav. 167:16-27. |
| Zhou FC et. al. (2016). Cell-Wide DNA De-Methylation and Re-Methylation of Purkinje Neurons in the Developing Cerebellum. PLoS One. 11(9):e0162063. |
| Gurioli G et. al. (2016). Methylation pattern analysis in prostate cancer tissue: identification of biomarkers using an MS-MLPA approach. J Transl Med. 14(1):249. |
| Zhang S et. al. (2016). Physiological and biochemical aspects of methionine isomers and a methionine analogue in broilers. Poult Sci. |
| Marandel L et. al. (2016). Remodelling of the hepatic epigenetic landscape of glucose-intolerant rainbow trout (Oncorhynchus mykiss) by nutritional status and dietary carbohydrates. Sci Rep. 6:32187. |
| Han B et. al. (2016). Variation of DNA Methylome of Zebrafish Cells under Cold Pressure. PLoS One. 11(8):e0160358. |
| Ganguly A et. al. (2016). Maternal Calorie Restriction Causing Utero-placental Insufficiency Differentially Affects Mammalian Placental Glucose and Leucine Transport Molecular Mechanisms. Endocrinology. :en20161259. |
| Putnam HM et. al. (2016). Ocean acidification influences host DNA methylation and phenotypic plasticity in environmentally susceptible corals Evol Appl. |
| Reichetzeder C et. al. (2016). Increased global placental DNA methylation levels are associated with gestational diabetes. Clin Epigenetics. 8:82. |
| Fang X et. al. (2016). Knockdown of DNA methyltransferase 3a alters gene expression and inhibits function of embryonic cardiomyocytes. FASEB J. |
| Larsson L et. al. (2016). Expression of TET2 enzyme indicates enhanced epigenetic modification of cells in periodontitis. Eur J Oral Sci. |
| Herencia C et. al. (2016). Procaine Inhibits Osteo/Odontogenesis through Wnt/β-Catenin Inactivation. PLoS One. 11(6):e0156788. |
| Jiang H et. al. (2016). Abnormal Epigenetic Modifications in Peripheral T Cells from Patients with Abdominal Aortic Aneurysm Are Correlated with Disease Development. J Vasc Res. 52(6):404-413. |
| de la Rocha C et. al. (2016). Associations between whole peripheral blood fatty acids and DNA methylation in humans. Sci Rep. 6:25867. |
| McDonald JI et. al. (2016). Reprogrammable CRISPR/Cas9-based system for inducing site-specific DNA methylation. Biol Open. |
| Pierce LM et. al. (2016). Long-Term Epigenetic Alterations in a Rat Model of Gulf War Illness. Neurotoxicology. |
| Kala R et. al. (2016). A Novel Combinatorial Epigenetic Therapy Using Resveratrol and Pterostilbene for Restoring Estrogen Receptor-α (ERα) Expression in ERα-Negative Breast Cancer Cells. PLoS One. 11(5):e0155057. |
| Navalta J et. al. (2016). The Epigenomic Marker Global DNA Methylation is Related to Measures of Body Composition and Aerobic Capacity in Females but not in Males Research Gate. |
| Çelik SU et. al. (2016). The sensitivity of 5-formylcytosine to doxorubicin regardless of DNA damage Turk J Biol. 40 |
| Zhong J et. al. (2016). The green tea polyphenol EGCG alleviates maternal diabetes-induced neural tube defects by inhibiting DNA hypermethylation. Am J Obstet Gynecol. |
| Almeida MR et. al. (2016). Maternal vitamin B6 deficient or supplemented diets on expression of genes related to GABAergic, serotonergic or glutamatergic pathways in hippocampus of rat dams and their offspring. Mol Nutr Food Res. |
| Liu Y et. al. (2016). Effects of Wutou Decoction on DNA Methylation and Histone Modifications in Rats with Collagen-Induced Arthritis Evid. Based Complement. Alternat. Med.. 2016:1-9. |
| Wijenayake S et. al. (2016). The role of DNA methylation during anoxia tolerance in a freshwater turtle (Trachemys scripta elegans). J Comp Physiol B. |
| Kima GH et. al. (2016). Cryptophyte gene regulation in the kleptoplastidic, karyokleptic ciliate Mesodinium rubrum Harmful Algae. 52:23-33. |
| Cui Y et. al. (2016). Epigenetic toxicity of trichloroethylene: a single-molecule perspective Toxicol Res. 5(2):641-650. |
| Aguilo F et. al. (2016). Deposition of 5-Methylcytosine on Enhancer RNAs Enables the Coactivator Function of PGC-1α. Cell Rep. 14(3):479-92. |
| Elliott E et. al. (2016). Dnmt3a in the Medial Prefrontal Cortex Regulates Anxiety-Like Behavior in Adult Mice. J Neurosci. 36(3):730-40. |
| Achille NJ et. al. (2016). Association between early promoter-specific DNA methylation changes and outcome in older acute myeloid leukemia patients. Leuk Res. |
| Kurdyukov S et. al. (2016). DNA Methylation Analysis: Choosing the Right Method. Biology (Basel). 5(1) |
| 2015年 |
| Mendoza-Perez J et. al. (2015). Genomic DNA hypomethylation and risk of renal cell carcinoma: A case-control study. Clin Cancer Res. |
| Sessions-Bresnahan DR et. al. (2015). Effect of Obesity on the Preovulatory Follicle and Lipid Fingerprint of Equine Oocytes. Biol Reprod. |
| Knorr E et. al. (2015). Translocation of bacteria from the gut to the eggs triggers maternal transgenerational immune priming in Tribolium castaneum. Biol Lett. 11(12) |
| Endo S et. al. (2015). Immunomodulatory drugs act as inhibitors of DNA methyltransferases and induce PU.1 up-regulation in myeloma cells Biochem Biophys Res Commun. |
| Sun Y et. al. (2015). DNA Methylation Modulates Nociceptive Sensitization after Incision. PLoS One. 10(11):e0142046. |
| Biergans SD et. al. (2015). Dnmts and Tet target memory-associated genes after appetitive olfactory training in honey bees. Sci Rep. 5:16223. |
| Morfouace M et. al. (2015). Preclinical studies of 5-fluoro-2'-deoxycytidine and tetrahydrouridine in pediatric brain tumors. J Neurooncol. |
| Liang J et. al. (2015). A correlation study on the effects of DNMT1 on methylation levels in CD4+ T cells of SLE patients. Int J Clin Exp Med. 8(10):19701-19708. |
| Patel S et. al. (2015). Establishment of Acute myeloid leukemic xenografts as preclinical models of epigenetic therapies Int J Hematol Res.. 1(3) |
| Hervouet E et. al. (2015). The autophagy GABARAPL1 gene is epigenetically regulated in breast cancer models. BMC Cancer. 15(1):729. |
| Song C et. al. (2015). Effect of the one‑carbon unit cycle on overall DNA methylation in children with Down's syndrome. Mol Med Rep. |
| Choudhury SR et. al. (2015). Selective increase in subtelomeric DNA methylation: an epigenetic biomarker for malignant glioma. Clin Epigenetics. 7:107. |
| Shin YH et. al. (2015). Epigenetic alteration of the purinergic type 7 receptor in salivary epithelial cells. Biochem Biophys Res Commun. |
| Steinritz D et. al. (2015). Epigenetic modulations in early endothelial cells and DNA hypermethylation in human skin after sulfur mustard exposure. Toxicol Lett. |
| Brown TA et. al. (2015). Alterations in DNA methylation corresponding with lung inflammation and as a biomarker for disease development after MWCNT exposure. Nanotoxicology. :1-9. |
| Yang M et. al. (2015). Epigenetic modulation of Chlorella (Chlorella vulgaris) on exposure to polycyclic aromatic hydrocarbons. Environ Toxicol Pharmacol. 40(3):758-763. |
| Granados ML et. al. (2015). Contributions of the Epidermal Growth Factor Receptor to Acquisition of Platinum Resistance in Ovarian Cancer Cells. PLoS One. 10(9):e0136893. |
| Philbrook NA et. al. (2015). Investigating the effects of in utero benzene exposure on epigenetic modifications in maternal and fetal CD-1 mice. Toxicol Appl Pharmacol. |
| Atilano SR et. al. (2015). Mitochondrial DNA variants can mediate methylation status of inflammation, angiogenesis and signaling genes. Hum Mol Genet. 24(16):4491-503. |
| Wood RK et. al. (2015). Developmental profiles and expression of the DNA methyltransferase genes in the fathead minnow (Pimephales promelas) following exposure to di-2-ethylhexyl phthalate. Fish Physiol Biochem. |
| Fang X et. al. (2015). Cyclic AMP induces hypertrophy and alters DNA methylation in HL-1 cardiomyocytes. Am J Physiol Cell Physiol. :ajpcell.00058.2015. |
| Lin AP et. al. (2015). D2HGDH regulates alpha-ketoglutarate levels and dioxygenase function by modulating IDH2. Nat Commun. 6:7768. |
| Yang X et. al. (2015). The Relationship Between Cognitive Impairment and Global DNA Methylation Decrease Among Aluminum Potroom Workers. J Occup Environ Med. 57(7):713-7. |
| Solís MT et. al. (2015). 5-azacytidine promotes microspore embryogenesis initiation by decreasing global DNA methylation, but prevents subsequent embryo development in rapeseed and barley. Front Plant Sci. 6:472. |
| Bose R et. al. (2015). Tet3 mediates stable glucocorticoid-induced alterations in DNA methylation and Dnmt3a/Dkk1 expression in neural progenitors. Cell Death Dis. 6:e1793. |
| Götze S et. al. (2015). Epigenetic Changes during Hepatic Stellate Cell Activation. PLoS One. 10(6):e0128745. |
| Chen Z et. al. (2015). Piwil1 causes epigenetic alteration of PTEN gene via upregulation of DNA methyltransferase in type I endometrial cancer. Biochem Biophys Res Commun. |
| Imanishi S et. al. (2015). Effect of combined deferasirox and 5-azacytidine treatment on human leukemia cells in vitro. Ann Hematol. |
| Kim SH et. al. (2015). Egg-specific expression of protein with DNA methyltransferase activity in the biocarcinogenic liver fluke Clonorchis sinensis. Parasitology. :1-11. |
| Lechat MM et. al. (2015). Seed response to strigolactone is controlled by abscisic acid-independent DNA methylation in the obligate root parasitic plant, Phelipanche ramosa L. Pomel. J Exp Bot. 66(11):3129-40. |
| Agarwal P et. al. (2015). CGGBP1 mitigates cytosine methylation at repetitive DNA sequences. BMC Genomics. 16:390. |
| Zembrzycki A et. al. (2015). Postmitotic regulation of sensory area patterning in the mammalian neocortex by Lhx2. Proc Natl Acad Sci U S A. |
| Chen X et. al. (2015). Analysis of DNA Methylation and Gene Expression in Radiation-Resistant Head and Neck Tumors. Epigenetics. |
| Arroyo-Jousse V et. al. (2015). Global DNA methylation and homocysteine levels are lower in type 1 diabetes patients Rev. méd. Chile. 143(5) |
| Zhong T et. al. (2015). Parental Neuropathic Pain Influences Emotion-Related Behavior in Offspring Through Maternal Feeding Associated with DNA Methylation of Amygdale in Rats. Neurochem Res. |
| Montrose L et. al. (2015). Evaluating the effect of ambient particulate pollution on DNA methylation in Alaskan sled dogs: Potential applications for a sentinel model of human health. Sci Total Environ. 512-513:489-94. |
| Mei Y et. al. (2015). Aging-associated formaldehyde-induced norepinephrine deficiency contributes to age-related memory decline. Aging Cell. |
| Omidvar V et. al. (2015). DNA Methylation and Transcriptomic Changes in Response to Different Lights and Stresses in 7B-1 Male-Sterile Tomato. PLoS One. 10(4):e0121864. |
| Wang Y et. al. (2015). Elevated tissue Cr levels, increased plasma oxidative markers, and global hypomethylation of blood DNA in male Sprague-Dawley rats exposed to potassium dichromate in drinking water. Environ Toxicol. |
| Cui Y et. al. (2015). Decitabine Nanoconjugate Sensitizes Human Glioblastoma Cells to Temozolomide. Mol Pharm. |
| Yang Y et. al. (2015). The effect of mycophenolic acid on epigenetic modifications in lupus CD4+T cells. Clin Immunol. |
| Aulakh SS et. al. (2015). Characterization of a potato activation-tagged mutant, nikku, and its partial revertant. Planta. |
| Cui Y et. al. (2015). Dissecting the behavior and function of MBD3 in DNA methylation homeostasis by single-molecule spectroscopy and microscopy. Nucleic Acids Res. |
| Bull C et. al. (2015). Cortisol Is Not Associated with Telomere Shortening or Chromosomal Instability in Human Lymphocytes Cultured under Low and High Folate Conditions. PLoS One. 10(3):e0119367. |
| Montjean D et. al. (2015). Sperm global DNA methylation level: association with semen parameters and genome integrity. Andrology. |
| Binder NK et. al. (2015). Male obesity is associated with changed spermatozoa Cox4i1 mRNA level and altered seminal vesicle fluid composition in a mouse model. Mol Hum Reprod. |
| Ge R et. al. (2015). DNA Methyl Transferase 1 Reduces Expression of SRD5A2 in the Aging Adult Prostate. Am J Pathol. 185(3):870-82. |
| Binder NK et. al. (2015). Paternal obesity in a rodent model affects placental gene expression in a sex-specific manner. Reproduction. |
| Li C et. al. (2015). DNA methylation and histone modification patterns during the late embryonic and early postnatal development of chickens. Poult Sci. |
| Lewinskaa A et. al. (2015). Capsaicin-induced genotoxic stress does not promote apoptosis in A549 human lung and DU145 prostate cancer cells. Mutation Res. 779:23-34. |
| Cleys ER et. al. (2015). Androgen receptor and histone lysine demethylases in ovine placenta. PLoS One. 10(2):e0117472. |
| Sun L et. al. (2015). MiR125a-5p acting as a novel Gab2 suppressor inhibits invasion of glioma. Mol Carcinog. |
| Wang P et. al. (2015). Aberrant hypermethylation of aldehyde dehydrogenase 2 promoter upstream sequence in rats with experimental myocardial infarction. Biomed Res Int. 2015:503692. |
| 2014年 |
| Arechederra M et. al. (2014). p38 MAPK down-regulates fibulin 3 expression through methylation of gene regulatory sequences. Role in migration and invasion. J Biol Chem. |
| Duan CG et. al. (2014). Specific but interdependent functions for Arabidopsis AGO4 and AGO6 in RNA-directed DNA methylation. EMBO J. |
| Fang X et. al. (2014). Caffeine exposure alters cardiac gene expression in embryonic cardiomyocytes. Am J Physiol Regul Integr Comp Physiol. 307(12):R1471-87. |
| Sim CB et. al. (2014). Dynamic changes in the cardiac methylome during postnatal development. FASEB J. |
| LePage DP et. al. (2014). The relative importance of DNA methylation and Dnmt2-mediated epigenetic regulation on Wolbachia densities and cytoplasmic incompatibility. PeerJ. 2:e678. |
| Nagre NN et. al. (2014). CB1-Receptor Knockout Neonatal Mice are Protected Against Ethanol-induced Impairments of DNMT1, DNMT3A and DNA Methylation. J Neurochem. |
| Shin YH et. al. (2014). Epigenetic modulation of the muscarinic type 3 receptor in salivary epithelial cells. Lab Invest. |
| Yang Q et. al. (2014). Epigenetic Features Induced by Ischemia-Hypoxia in Cultured Rat Astrocytes. Mol Neurobiol. |
| Freetly HC et. al. (2014). The consequence of level of nutrition on heifer ovarian and mammary development. J Anim Sci. 92(12):5437-43. |
| Li C et. al. (2014). Maternal high-zinc diet attenuates intestinal inflammation by reducing DNA methylation and elevating H3K9 acetylation in the A20 promoter of offspring chicks. J Nutr Biochem. |
| Leo E et. al. (2014). DNA Methyltransferase 1 Drives Transcriptional Down-Modulation of β Catenin Antagonist Chibby1 Associated With the BCR-ABL1 Gene of Chronic Myeloid Leukemia. J Cell Biochem. |
| Puig-Vilanova E et. al. (2014). Epigenetic mechanisms in respiratory muscle dysfunction of patients with chronic obstructive pulmonary disease. PLoS One. 9(11):e111514. |
| Ward A et. al. (2014). The deregulated promoter methylation of the Polo-like kinases as a potential biomarker in hematological malignancies. Leuk Lymphoma. :1-25. |
| Murphy TM et. al. (2014). Anxiety is associated with higher levels of global DNA methylation and altered expression of epigenetic and interleukin-6 genes. Psychiatr Genet. |
| Lopez-Bertoni H et. al. (2014). DNMT-dependent suppression of microRNA regulates the induction of GBM tumor-propagating phenotype by Oct4 and Sox2. Oncogene. |
| van Esterik JC et. al. (2014). Liver DNA methylation analysis in adult female C57BL/6JxFVB mice following perinatal exposure to bisphenol A. Toxicol Lett. 232(1):293-300. |
| Jung SH et. al. (2014). Molecular mechanisms of repeated social defeat-induced glucocorticoid resistance: Role of microRNA. Brain Behav Immun. |
| Wojtasik W et. al. (2014). Oligonucleotide treatment causes flax ß-glucanase up-regulation via changes in gene-body methylation. BMC Plant Biol. 14(1):261. |
| Biggar Y et. al. (2014). Global DNA modifications suppress transcription in brown adipose tissue during hibernation. Cryobiology. 69(2):333-8. |
| Lee Y et. al. (2014). Radiation-induced changes in DNA methylation and their relationship to chromosome aberrations in nuclear power plant workers. Int J Radiat Biol. :1-32. |
| Brown TC et. al. (2014). Frequent Silencing of RASSF1A via Promoter Methylation in Follicular Thyroid Hyperplasia: A Potential Early Epigenetic Susceptibility Event in Thyroid Carcinogenesis. JAMA Surg. |
| Petr Cápal et. al. (2014). Expression and epigenetic profile of protoplast cultures (Cucumis sativus L.) In Vitro Cellular & Developmental Biology - Plant. |
| Lashley T et. al. (2014). Alterations in global DNA methylation and hydroxymethylation are not detected in Alzheimer's disease. Neuropathol Appl Neurobiol. |
| Song Y et. al. (2014). Transgenerational impaired male fertility with an Igf2 epigenetic defect in the rat are induced by the endocrine disruptor, p,p'-DDE. Hum Reprod. |
| Rodríguez-Sanz H et. al. (2014). Early markers are present in both embryogenesis pathways from microspores and immature zygotic embryos in cork oak, Quercus suber L. BMC Plant Biol. 14(1):224. |
| Kim SE et. al. (2014). One-pot approach for examining the DNA methylation patterns using an engineered methyl-probe. Biosens Bioelectron. 58:333-7. |
| Naito T et. al. (2014). IGF2 differentially methylated region hypomethylation in relation to pathological and molecular features of serrated lesions. World J Gastroenterol. 20(29):10050-61. |
| Zhou Y et. al. (2014). Genome-Wide Demethylation by 5-aza-2'-Deoxycytidine Alters the Cell Fate of Stem/Progenitor Cells. Stem Cell Rev. |
| Jin L et. al. (2014). Genome-wide DNA methylation changes in skeletal muscle between young and middle-aged pigs. BMC Genomics. 15(1):653. |
| Liu Y et. al. (2014). Global and cyp19a1a gene specific DNA methylation in gonads of adult rare minnow Gobiocypris rarus under bisphenol A exposure. Aquat Toxicol. 156C:10-16. |
| Lin JR et. al. (2014). Vitamin C Protects Against UV Irradiation-Induced Apoptosis Through Reactivating Silenced Tumor Suppressor Genes p21 and p16 in a Tet-Dependent DNA Demethylation Manner in Human Skin Cancer Cells. Cancer Biother Radiopharm. 29(6):257-64. |
| El-Tantawy AA et. al. (2014). Changes in DNA methylation levels and nuclear distribution patterns after microspore reprogramming to embryogenesis in barley. Cytogenet Genome Res. 143(1-3):200-8. |
| Marino AM et. al. (2014). Effects of epigenetic modificators in combination with small molecule inhibitors of receptor tyrosine kinases on medulloblastoma growth. Biochem Biophys Res Commun. |
| Puig-Vilanova E et. al. (2014). Do epigenetic events take place in the vastus lateralis of patients with mild chronic obstructive pulmonary disease? PLoS One. 9(7):e102296. |
| Zhang H et. al. (2014). Maternal vitamin D deficiency during pregnancy results in insulin resistance in rat offspring, which is associated with inflammation and Iκbα methylation. Diabetologia. |
| Katz TA et. al. (2014). Inhibition of histone demethylase, LSD2 (KDM1B), attenuates DNA methylation and increases sensitivity to DNMT inhibitor-induced apoptosis in breast cancer cells. Breast Cancer Res Treat. 146(1):99-108. |
| Putiri EL et. al. (2014). Distinct and overlapping control of 5-methylcytosine and 5-hydroxymethylcytosine by the TET proteins in human cancer cells. Genome Biol. 15(6):R81. |
| Götze S et. al. (2014). Epigenetic regulation during hepatic stellate cell activation. Eur J Med Res. 19(Suppl 1):S7. |
| Conti Ad et. al. (2014). Dose- and time-dependent epigenetic changes in the livers of fisher 344 rats exposed to furan. Toxicol Sci. 139(2):371-80. |
| Bujko M et. al. (2014). Repetitive genomic elements and overall DNA methylation changes in acute myeloid and childhood B-cell lymphoblastic leukemia patients. Int J Hematol. |
| Gaglio D et. al. (2014). Learning induced epigenetic modifications in the ventral striatum are necessary for long-term memory. Behav Brain Res. 265:61-8. |
| Saldanha SN et. al. (2014). Molecular mechanisms for inhibition of colon cancer cells by combined epigenetic-modulating epigallocatechin gallate and sodium butyrate. Exp Cell Res. 324(1):40-53. |
| Calvo X et. al. (2014). High levels of global DNA methylation are an independent adverse prognostic factor in a series of 90 patients with de novo myelodysplastic syndrome. Leuk Res. |
| So MY et. al. (2014). Gene expression profile and toxic effects in human bronchial epithelial cells exposed to zearalenone. PLoS One. 9(5):e96404. |
| Khalid O et. al. (2014). Gene expression signatures affected by alcohol-induced DNA methylomic deregulation in human embryonic stem cells. Stem Cell Res. 12(3):791-806. |
| Miousse IR et. al. (2014). Epigenetic alterations induced by ambient particulate matter in mouse macrophages. Environ Mol Mutagen. 55(5):428-35. |
| Trivedi M et. al. (2014). Morphine induces redox-based changes in global DNA methylation and retrotransposon transcription by inhibition of excitatory amino acid transporter type 3-mediated cysteine uptake. Mol Pharmacol. 85(5):747-57. |
| Klengel T et. al. (2014). The role of DNA methylation in stress-related psychiatric disorders. Neuropharmacology. 80:115-32. |
| Tellez-Plaza M et. al. (2014). Association of Global DNA Methylation and Global DNA Hydroxymethylation with Metals and other Exposures in Human Blood DNA Samples. Environ Health Perspect. |
| Hadoux J et. al. (2014). SDHB mutations are associated with response to temozolomide in patients with metastatic pheochromocytoma or paraganglioma. Int J Cancer. |
| Basu A et. al. (2014). The CpG Island Encompassing the Promoter and First Exon of Human DNMT3L Gene Is a PcG/TrX Response Element (PRE). PLoS One. 9(4):e93561. |
| Kang KA et. al. (2014). Epigenetic modification of Nrf2 in 5-fluorouracil-resistant colon cancer cells: involvement of TET-dependent DNA demethylation. Cell Death Dis. 5:e1183. |
| Yan H et. al. (2014). piRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia. |
| Huang Y et. al. (2014). Maternal high folic acid supplement promotes glucose intolerance and insulin resistance in male mouse offspring fed a high-fat diet. Int J Mol Sci. 15(4):6298-313. |
| Wu X et. al. (2014). Epigenetic Signature of Chronic Cerebral Hypoperfusion and Beneficial Effects of S-adenosylmethionine in Rats. Mol Neurobiol. |
| Dhliwayo N et. al. (2014). Parp inhibition prevents ten eleven translocase enzyme activation and hyperglycemia induced DNA demethylation. Diabetes. |
| Batra V et. al. (2014). Dietary l-methionine supplementation mitigates gamma-radiation induced global DNA hypomethylation: Enhanced metabolic flux towards S-adenosyl-l-methionine (SAM) biosynthesis increases genomic methylation potential. Food Chem Toxicol. 69C:46-54. |
| Nosho K et. al. (2014). Association of microRNA-31 with BRAF mutation, colorectal cancer survival and serrated pathway. Carcinogenesis. 35(4):776-83. |
| Siuda D et. al. (2014). Social isolation-induced epigenetic changes in midbrain of adult mice. J Physiol Pharmacol. 65(2):247-55. |
| Huzayyin AA et. al. (2014). Decreased global methylation in patients with bipolar disorder who respond to lithium. Int J Neuropsychopharmacol. 17(4):561-9. |
| Wu J et. al. (2014). Kaposi's sarcoma-associated herpesvirus (KSHV) vIL-6 promotes cell proliferation and migration by upregulating DNMT1 via STAT3 activation. PLoS One. 9(3):e93478. |
| Wang LJ et. al. (2014). Betaine attenuates hepatic steatosis by reducing methylation of the MTTP promoter and elevating genomic methylation in mice fed a high-fat diet. J Nutr Biochem. 25(3):329-36. |
| Azzi A et. al. (2014). Circadian behavior is light-reprogrammed by plastic DNA methylation. Nat Neurosci. 17(3):377-82. |
| Ufkin ML et. al. (2014). miR-125a regulates cell cycle, proliferation, and apoptosis by targeting the ErbB pathway in acute myeloid leukemia. Leuk Res. 38(3):402-10. |
| Cho Y et. al. (2014). Colon cancer cell apoptosis is induced by combined exposure to the n-3 fatty acid docosahexaenoic acid and butyrate through promoter methylation. Exp Biol Med (Maywood). 239(3):302-10. |
| Anier K et. al. (2014). Maternal separation is associated with DNA methylation and behavioural changes in adult rats. Eur Neuropsychopharmacol. 24(3):459-68. |
| Saheb A et. al. (2014). Probing for DNA methylation with a voltammetric DNA detector. Analyst. 139(4):786-92. |
| Weng X et. al. (2014). DNA methylation profiling in the thalamus and hippocampus of postnatal malnourished mice, including effects related to long-term potentiation. BMC Neurosci. 15:31. |
| Kristensen DG et. al. (2014). Evidence that active demethylation mechanisms maintain the genome of carcinoma in situ cells hypomethylated in the adult testis. Br J Cancer. 110(3):668-78. |
| Xu Q et. al. (2014). DNA methylation and regulation of the CD8A after duck hepatitis virus type 1 infection. PLoS One. 9(2):e88023. |
| Zhu JQ et. al. (2014). Sodium fluoride disrupts DNA methylation of H19 and Peg3 imprinted genes during the early development of mouse embryo. Arch Toxicol. 88(2):241-8. |
| Kim M et. al. (2014). Dynamic changes in DNA methylation and hydroxymethylation when hES cells undergo differentiation toward a neuronal lineage. Hum Mol Genet. 23(3):657-67. |
| Guo F et. al. (2014). Alternative splicing, promoter methylation, and functional SNPs of sperm flagella 2 gene in testis and mature spermatozoa of Holstein bulls. Reproduction. 147(2):241-52. |
| Chaparro et. al. (2014). P651 Identification of new genetic variants related to thiopurine-induced myelotoxicity in inflammatory bowel disease (IBD) patients with normal thiopurines-methyltransferase (TPMT): a genome-wide association study J Crohns Colitis. 8(1) |
| Ward A et. al. (2014). p53-Dependent and cell specific epigenetic regulation of the polo-like kinases under oxidative stress. PLoS One. 9(1):e87918. |
| Kehrmann J et. al. (2014). Impact of 5-Aza-2`-deoxycytidine and Epigallocatechin-3-gallate for induction of human regulatory T cells. Immunology. |
| Buscariollo DL et. al. (2014). Embryonic caffeine exposure acts via A1 adenosine receptors to alter adult cardiac function and DNA methylation in mice. PLoS One. 9(1):e87547. |
| Weiner AS et. al. (2014). Methylenetetrahydrofolate reductase C677T and methionine synthase A2756G polymorphisms influence on leukocyte genomic DNA methylation level. Gene. 533(1):168-72. |
| 2013年 |
| Nettersheim D et. al. (2013). Analysis of TET expression/activity and 5mC oxidation during normal and malignant germ cell development. PLoS One. 8(12):e82881. |
| Senesi P et. al. (2013). DNA demethylation enhances myoblasts hypertrophy during the late phase of myogenesis activating the IGF-I pathway. Endocrine. |
| Wu X et. al. (2013). Chronic cerebrovascular hypoperfusion affects global DNA methylation and histone acetylation in rat brain. Neurosci Bull. 29(6):685-92. |
| Falk IJ et. al. (2013). Decreased survival in normal karyotype AML with single-nucleotide polymorphisms in genes encoding the AraC metabolizing enzymes cytidine deaminase and 5'-nucleotidase. Am J Hematol. 88(12):1001-6. |
| Nagaraju GP et. al. (2013). Novel synthetic curcumin analogues EF31 and UBS109 are potent DNA hypomethylating agents in pancreatic cancer. Cancer Lett. 341(2):195-203. |
| Kim JE et. al. (2013). Characterization of non-CG genomic hypomethylation associated with gamma-ray-induced suppression of CMT3 transcription in Arabidopsis thaliana. Radiat Res. 180(6):638-48. |
| Moldrich RX et. al. (2013). Inhibition of histone deacetylase in utero causes sociability deficits in postnatal mice. Behav Brain Res. 257:253-64. |
| Moscariello M et. al. (2013). Effects of chromatin decondensation on alternative NHEJ. DNA Repair (Amst). 12(11):972-81. |
| Zhao HC et. al. (2013). Aberrant epigenetic modification in murine brain tissues of offspring from preimplantation genetic diagnosis blastomere biopsies. Biol Reprod. 89(5):117. |
| Kittan NA et. al. (2013). Cytokine induced phenotypic and epigenetic signatures are key to establishing specific macrophage phenotypes. PLoS One. 8(10):e78045. |
| Simmons RK et. al. (2013). DNA methylation markers in the postnatal developing rat brain. Brain Res. 1533:26-36. |
| Jenkins TG et. al. (2013). Paternal aging and associated intraindividual alterations of global sperm 5-methylcytosine and 5-hydroxymethylcytosine levels. Fertil Steril. 100(4):945-51. |
| Bradley-Whitman MA et. al. (2013). Epigenetic changes in the progression of Alzheimer's disease. Mech Ageing Dev. 134(10):486-95. |
| Dror N et. al. (2013). Unique anti-glioblastoma activities of hypericin are at the crossroad of biochemical and epigenetic events and culminate in tumor cell differentiation. PLoS One. 8(9):e73625. |
| Testillano PS et. al. (2013). The 5-methyl-deoxy-cytidine (5mdC) localization to reveal in situ the dynamics of DNA methylation chromatin pattern in a variety of plant organ and tissue cells during development. Physiol Plant. 149(1):104-13. |
| Marquardt JU et. al. (2013). Sirtuin-6-dependent genetic and epigenetic alterations are associated with poor clinical outcome in hepatocellular carcinoma patients. Hepatology. 58(3):1054-64. |
| Fang X et. al. (2013). Global and gene specific DNA methylation changes during zebrafish development. Comp Biochem Physiol B Biochem Mol Biol. 166(1):99-108. |
| Sung-Hwa Sohn et. al. (2013). Screening for herbal medicines that affect ZIC1 gene methylation in colorectal cancer Molecular & Cellular Toxicology. 9(3):211-218. Abstract |
| Lou J et. al. (2013). Role of DNA methylation in cell cycle arrest induced by Cr (VI) in two cell lines. PLoS One. 8(8):e71031. |
| O'Brien E et. al. (2013). Perinatal bisphenol A exposures increase production of pro-inflammatory mediators in bone marrow-derived mast cells of adult mice. J Immunotoxicol. |
| Shang Y et. al. (2013). Epigenetic alterations by DNA methylation in house dust mite-induced airway hyperresponsiveness. Am J Respir Cell Mol Biol. 49(2):279-87. |
| Williams-Karnesky RL et. al. (2013). Epigenetic changes induced by adenosine augmentation therapy prevent epileptogenesis. J Clin Invest. 123(8):3552-63. |
| Mejos KK et. al. (2013). Effects of parental folate deficiency on the folate content, global DNA methylation, and expressions of FRα, IGF-2 and IGF-1R in the postnatal rat liver. Nutr Res Pract. 7(4):281-6. |
| Sunahori K et. al. (2013). The catalytic subunit of protein phosphatase 2A (PP2Ac) promotes DNA hypomethylation by suppressing the phosphorylated mitogen-activated protein kinase/extracellular signal-regulated kinase (ERK) kinase (MEK)/phosphorylated ERK/DNMT1 protein pathway in T-cells from controls and systemic lupus erythematosus patients. J Biol Chem. 288(30):21936-44. |
| Ryley Parrish R et. al. (2013). Status epilepticus triggers early and late alterations in brain-derived neurotrophic factor and NMDA glutamate receptor Grin2b DNA methylation levels in the hippocampus. Neuroscience. 248C:602-619. |
| Kingham E et. al. (2013). Nanotopographical cues augment mesenchymal differentiation of human embryonic stem cells. Small. 9(12):2140-51. |
| Blossom SJ et. al. (2013). Metabolic changes and DNA hypomethylation in cerebellum are associated with behavioral alterations in mice exposed to trichloroethylene postnatally. Toxicol Appl Pharmacol. 269(3):263-9. |
| Fernández de Larrea C et. al. (2013). Impact of global and gene-specific DNA methylation pattern in relapsed multiple myeloma patients treated with bortezomib. Leuk Res. 37(6):641-6. |
| Mukhopadhyay P et. al. (2013). Alcohol modulates expression of DNA methyltranferases and methyl CpG-/CpG domain-binding proteins in murine embryonic fibroblasts. Reprod Toxicol. 37:40-8. |
| Qin HH et. al. (2013). Associations between aberrant DNA methylation and transcript levels of DNMT1 and MBD2 in CD4+T cells from patients with systemic lupus erythematosus. Australas J Dermatol. 54(2):90-5. |
| Patel BB et. al. (2013). Lifelong exposure to bisphenol a alters cardiac structure/function, protein expression, and DNA methylation in adult mice. Toxicol Sci. 133(1):174-85. |
| Kim KI et. al. (2013). Changes in the epigenetic status of the SOX-9 promoter in human osteoarthritic cartilage. J Bone Miner Res. 28(5):1050-60. |
| Kim HW et. al. (2013). Effects of paternal folate deficiency on the expression of insulin-like growth factor-2 and global DNA methylation in the fetal brain. Mol Nutr Food Res. 57(4):671-6. |
| Delaney C et. al. (2013). Maternal diet supplemented with methyl-donors protects against atherosclerosis in F1 ApoE(-/-) mice. PLoS One. 8(2):e56253. |
| Craig PM et. al. (2013). Methionine restriction affects the phenotypic and transcriptional response of rainbow trout (Oncorhynchus mykiss) to carbohydrate-enriched diets. Br J Nutr. 109(3):402-12. |
| Wiley KL et. al. (2013). Ethnic differences in DNA methyltransferases expression in patients with systemic lupus erythematosus. J Clin Immunol. 33(2):342-8. |
| Murphy TM et. al. (2013). Genetic variation in DNMT3B and increased global DNA methylation is associated with suicide attempts in psychiatric patients. Genes Brain Behav. 12(1):125-32. |
| Word B et. al. (2013). Cigarette smoke condensate induces differential expression and promoter methylation profiles of critical genes involved in lung cancer in NL-20 lung cells in vitro: short-term and chronic exposure. Int J Toxicol. 32(1):23-31. |
| Nischal S et. al. (2013). Methylome profiling reveals distinct alterations in phenotypic and mutational subgroups of myeloproliferative neoplasms. Cancer Res. 73(3):1076-85. |
| Chai J et. al. (2013). Molecular cloning, tissue expression, and analysis with genome DNA methylation of porcine LSD1 gene. Appl Biochem Biotechnol. 169(1):134-44. |
| Qin H et. al. (2013). MicroRNA-29b contributes to DNA hypomethylation of CD4+ T cells in systemic lupus erythematosus by indirectly targeting DNA methyltransferase 1. J Dermatol Sci. 69(1):61-7. |
| Hodgson N et. al. (2013). Soluble oligomers of amyloid-β cause changes in redox state, DNA methylation, and gene transcription by inhibiting EAAT3 mediated cysteine uptake. J Alzheimers Dis. 36(1):197-209. |
| 2012年 |
| Figueroa-Romero C et. al. (2012). Identification of epigenetically altered genes in sporadic amyotrophic lateral sclerosis. PLoS One. 7(12):e52672. |
| Palacios JA et. al. (2012). Long-term nonprogressor and elite controller patients who control viremia have a higher percentage of methylation in their HIV-1 proviral promoters than aviremic patients receiving highly active antiretroviral therapy. J Virol. 86(23):13081-4. |
| Bose R et. al. (2012). Inherited effects of low-dose exposure to methylmercury in neural stem cells. Toxicol Sci. 130(2):383-90. |
| Arakawa Y et. al. (2012). Association of polymorphisms in DNMT1, DNMT3A, DNMT3B, MTHFR and MTRR genes with global DNA methylation levels and prognosis of autoimmune thyroid disease. Clin Exp Immunol. 170(2):194-201. |
| Rehan VK et. al. (2012). Perinatal nicotine exposure induces asthma in second generation offspring. BMC Med. 10:129. |
| Huang SK et. al. (2012). Prostaglandin E₂ increases fibroblast gene-specific and global DNA methylation via increased DNA methyltransferase expression. FASEB J. 26(9):3703-14. |
| Govorko D et. al. (2012). Male germline transmits fetal alcohol adverse effect on hypothalamic proopiomelanocortin gene across generations. Biol Psychiatry. 72(5):378-88. |
| Yamamura Y et. al. (2012). DNA demethylating agent decitabine increases AQP5 expression and restores salivary function. J Dent Res. 91(6):612-7. |
| Fernàndez-Roig S et. al. (2012). Vitamin B12 deficiency in the brain leads to DNA hypomethylation in the TCblR/CD320 knockout mouse. Nutr Metab (Lond). 9:41. |
| Poloni A et. al. (2012). Human dedifferentiated adipocytes show similar properties to bone marrow-derived mesenchymal stem cells. Stem Cells. 30(5):965-74. |
| Gilbert KM et. al. (2012). Epigenetic alterations regulate temporary reversal of CD4(+) T cell activation caused by trichloroethylene exposure. Toxicol Sci. 127(1):169-78. |
| Wang TC et. al. (2012). Oxidative DNA damage and global DNA hypomethylation are related to folate deficiency in chromate manufacturing workers. J Hazard Mater. 213-214:440-6. |
| Chen H et. al. (2012). Effect of aging on 5-hydroxymethylcytosine in the mouse hippocampus. Restor Neurol Neurosci. 30(3):237-45. |
| Siddiqi S et. al. (2012). Hiwi mediated tumorigenesis is associated with DNA hypermethylation. PLoS One. 7(3):e33711. |
| 2011年 |
| Hsu A et. al. (2011). Promoter de-methylation of cyclin D2 by sulforaphane in prostate cancer cells. Clin Epigenetics. 3:3. |
| Li W et. al. (2011). Distribution of 5-hydroxymethylcytosine in different human tissues. J Nucleic Acids. 2011:870726. |
| Ward A et. al. (2011). Aberrant methylation of Polo-like kinase CpG islands in Plk4 heterozygous mice. BMC Cancer. 11:71. |
追加しました。
製品情報は掲載時点のものですが、価格表内の価格については随時最新のものに更新されます。お問い合わせいただくタイミングにより製品情報・価格などは変更されている場合があります。
表示価格に、消費税等は含まれていません。一部価格が予告なく変更される場合がありますので、あらかじめご了承下さい。