PNGase F PRIME™ Glycosidase
Date:July 10 2020Web Page No:81437

* This product can be exported for South Korea and Taiwan customers.
PNGase F PRIME™ Glycosidase is a mutant recombinant PNGase F, cloned from Flavobacterium meningosepticum and expressed and purified from E. coli.
PNGase F PRIME™ Glycosidase is shown to have better characteristics, compared to other commercially available PNGase F - for example, PNGase F PRIME™ Glycosidase can remove N-glycan which can not be accomplised by wild type PNGase F.
This unique character enables to analyze sugar chain structure which could not be analyzed by mass spectrometry until now.
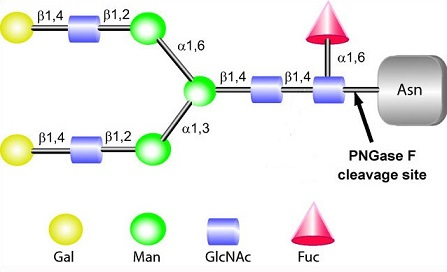
Features
- Wider specificity to N-glycan than conventional PNGase F.
- Can check easily if the target proteins have N-glycan modifcation or not.
- Unit of enzyme will completely deglycosylate 10 μg of denatured RNase B following incubation for 10 minutes at 37℃.
- High S/N ratio
- Lyophilized type (PNGase F PRIME-LY) is also available
Example Data
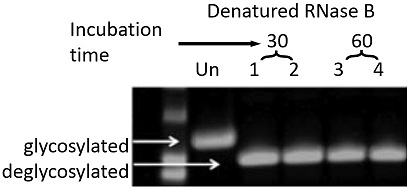
Fig.1 RNase B was treated by PNGase F PRIME™ Glycosidase for 30 min (Lane 1, 2) or 60 min (Lane 3, 4) and denatured. Following after electrophoresis, proteins were stained by coomassie dyes to see deglycosylation.
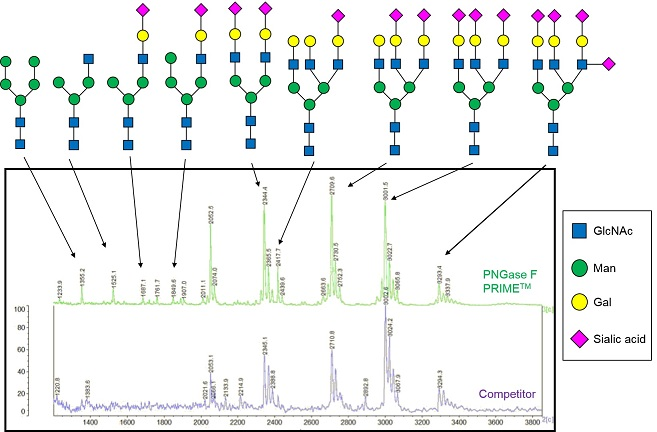
Fig.2 Anti-VEGF antibody (IgG1) was treated by PNGase F PRIME™ Glycosidase or other vendor's PNGase F and analyzed by HPLC.
PNGase F PRIME™ Glycosidase detects total 11 sugar chains, which include the ones traditional PNGase F could not remove.
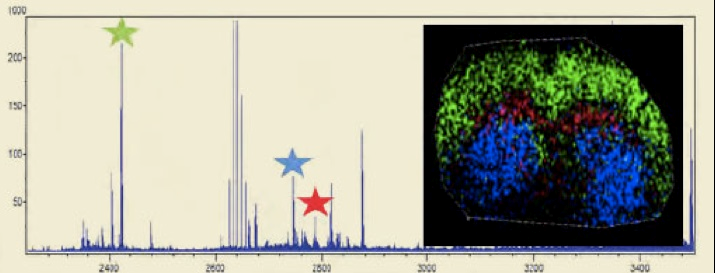
Fig. 3 MALDI-IMS Analysis of a Mouse Brain Slice.
The three starred oligosaccharide groups in the mass spec trace above are color coded and overlaid on a mouse brain section. The distribution shows a distinct localization pattern.
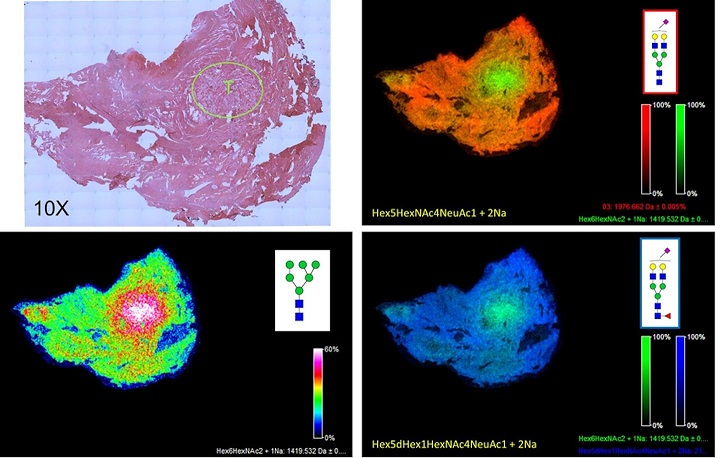
Fig.4 N-glycan imaging of a prostate cancer FFPE tissue.
A 5 micron FFPE slice of a human prostate cancer (Gleason 7) was antigen retrieved and incubated with PNGase F PRIME™ as described in Powers et al.
Images were acquired using a 7T MALDI-FTICR Solarix mass spectrometer and FlexImaging 4.1 software.
Shown in the upper right panel is a H&E stained image of the tissue (at 10X magnification), with the area of the highest density of tumor shown in the circle, marked with a T.
The bottom left panel shows a typical tumor N-glycan pattern for a single glycan, an Man6 at m/z=1419.
In the top right panel and bottom right panel, a two-glycan overlay is shown for the tumor glycan at m/z = 1419 (in green) and a mono-sialylated biantennary glycan at m/z = 1976 (top right panel, orange) and a mono-sialylated and core fucosylated glycan at m/z = 2122 (bottom right panel, blue).
Publications
(2014)
MALDI Imaging Mass Spectrometry Profiling of N-Glycans in Formalin-Fixed Paraffin Embedded Clinical Tissue Blocks and Tissue Microarrays.
PLoS One. 9(9):e106255
Powers TW, Neely BA, Shao Y, Tang H, Troyer DA, Mehta AS, Haab BB, Drake RR.
(2015)
Two-Dimensional N-Glycan Distribution Mapping of Hepatocellular Carcinoma Tissues by MALDI-Imaging Mass Spectrometry.
Biomolecules, 5, 2554-2572.
Powers, T.W. Holst, S., Wuhrer, M.,Mehta, A.S., Drake, R.R.
(2016)
Multimodal Mass Spectrometry Imaging of N-Glycans and Proteins from the Same Tissue Section.
Heijs B, Holst S, Briaire-de Bruijn IH, van Pelt GW, de Ru AH, van Veelen PA, Drake RR, Mehta AS, Mesker WE, Tollenaar RA, Bovée JV, Wuhrer M, McDonnell LA.
Anal Chem. 2016 Aug 2;88(15):7745-53. doi: 10.1021/acs.analchem.6b01739. Epub 2016 Jul 14.
Linkage-Specific in Situ Sialic Acid Derivatization for N-Glycan Mass Spectrometry Imaging of Formalin-Fixed Paraffin-Embedded Tissues.
Holst S, Heijs B, de Haan N, van Zeijl RJ, Briaire-de Bruijn IH, van Pelt GW, Mehta AS, Angel PM, Mesker WE, Tollenaar RA, Drake RR, Bovée JV, McDonnell LA, Wuhrer M.
Anal Chem. 2016 Jun 7;88(11):5904-13. doi: 10.1021/acs.analchem.6b00819. Epub 2016 May 23.
Glycosylation pattern of anti-platelet IgG is stable during pregnancy and predicts clinical outcome in alloimmune thrombocytopenia.
Sonneveld ME, Natunen S, Sainio S, Koeleman CA, Holst S, Dekkers G, Koelewijn J, Partanen J, van der Schoot CE, Wuhrer M, Vidarsson G.
Br J Haematol. 2016 Jul;174(2):310-20. doi: 10.1111/bjh.14053. Epub 2016 Mar 28.
N-glycosylation Profiling of Colorectal Cancer Cell Lines Reveals Association of Fucosylation with Differentiation and Caudal Type Homebox 1 (CDX1)/Villin mRNA Expression.
Holst S, Deuss AJ, van Pelt GW, van Vliet SJ, Garcia-Vallejo JJ, Koeleman CA, Deelder AM, Mesker WE, Tollenaar RA, Rombouts Y, Wuhrer M.
Mol Cell Proteomics. 2016 Jan;15(1):124-40. doi: 10.1074/mcp.M115.051235. Epub 2015 Nov
(2017)
High-Throughput and High-Sensitivity Mass Spectrometry-Based N-Glycomics of Mammalian Cells.
Holst S, van Pelt GW, Mesker WE, Tollenaar RA, Belo AI, van Die I, Rombouts Y, Wuhrer M.
Methods Mol Biol. 2017;1503:185-196.
MALDI Mass Spectrometry Imaging of N-Linked Glycans in Cancer Tissues.
Drake RR, Powers TW, Jones EE, Bruner E, Mehta AS, Angel PM.
Adv Cancer Res. 2017;134:85-116. doi: 10.1016/bs.acr.2016.11.009. Epub 2016 Dec 20. Review
(2018)
Human Disease Glycomics: Technology Advances Enabling Protein Glycosylation Analysis - Part 1.
Everest-Dass AV, Moh ESX, Ashwood C, Shathili AMM, Packer NH.
Expert Rev Proteomics: 2018 Feb;15(2):165-182. doi: 10.1080/14789450.2018.1421946. Epub 2018 Jan 11.
Human soluble phospholipase A2 receptor is an inhibitor of the integrin-mediated cell migratory response to collagen-I.
Watanabe K, Watanabe K, Watanabe Y, Fujioka D, Nakamura T, Nakamura K, Obata JE, Kugiyama K.
Am J Physiol Cell Physiol. 2018 Sep 1;315(3):C398-C408.
doi: 10.1152/ajpcell.00239.2017. Epub 2018 May 23.
(2019)
Antibody panel based N-glycan imaging for N-glycoprotein biomarker discovery.
Black, AP, Angel, PM, Drake, RR, Mehta, AS.
Current Protocols in Protein Science, (2019) 98, e99. doi: 10.1002/cpps.99
Cites the use of PNGase F PRIME™ in multiplexed analysis of N-glycoproteins from biofluids.
Carbohydrate-binding property of a cell wall integrity and stress response component (WSC) domain of an alcohol oxidase from the rice blast pathogen Pyricularia oryzae.
Oide S, Tanaka Y, Watanabe A, Inui M.
Enzyme Microbial Technology. 2019 Jun;125:13-20, doi:10.1016/j.enzmictec.2019.02.009. Epub 2019 Feb 25.
Cites the use of PNGase F PRIME™ in TritonX-100 nonionic detergent.
Defining the human kidney N-glycome in normal and cancer tissues using MALDI imaging mass spectrometry.
Drake RR, McDowell C, West C, David F, Powers TW, Nowling T, Bruner E, Mehta AS, Angel PM, Marlow LA, Tun HW, Copland JA.
J Mass Spectrom. 2019 Dec 20.
(2020)
How presence of a signal peptide affects human galectins-1 and -4: Clues to explain common absence of a leader sequence among adhesion/growth-regulatory galectins.
Kutzner TJ, Higuero AM, Süßmair M, Kopitz J, Hingar M, Díez-Revuelta N, Caballero GG, Kaltner H, Lindner I, Abad-Rodríguez J, Reusch D, Gabius HJ.
Biochim Biophys Acta Gen Subj. 2020 Jan;1864(1):129449.
[Date : March 10 2026 00:08]
| Detail | Product Name | Product Code | Supplier | Size | Price | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
PNGase F PRIME (TM) DatasheetThis may not be the latest data sheet. |
NZS1 | NZS | 50000 units | - | |||||||||||||||||||||||||||||||
|
|
|
||||||||||||||||||||||||||||||||||
|
PNGase F PRIME-LY DatasheetThis may not be the latest data sheet. |
NZS1-LY | NZS | 50000 units | - | |||||||||||||||||||||||||||||||
|
|
|
||||||||||||||||||||||||||||||||||
[Date : March 10 2026 00:08]
PNGase F PRIME (TM)
DatasheetThis may not be the latest data sheet.
- Product Code: NZS1
- Supplier: NZS
- Size: 50000units
- Price: -
| Description |
Only available for South Korea and Taiwan customers. |
||
|---|---|---|---|
| Storage | -20°C | CAS | |
| Link |
|
||
PNGase F PRIME-LY
DatasheetThis may not be the latest data sheet.
- Product Code: NZS1-LY
- Supplier: NZS
- Size: 50000units
- Price: -
| Description |
Only available for South Korea and Taiwan customers. |
||
|---|---|---|---|
| Storage | RT,Dry | CAS | |
| Link |
|
||
CONTACT
export@funakoshi.co.jp
- ※Prices on our website are for your reference only. Please inquire your distributor for your prices.
- ※Please note that Product Information or Price may change without notice.