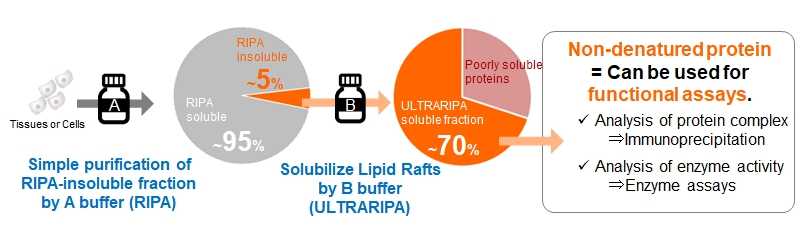
You can extract lipid raft enriched proteins by combining two kinds of buffers UltraRIPA kit for Lipid Raft
Date:November 14 2017Web Page No:80549
This buffer kit enables rapid and efficient extraction of lipid raft proteins that are difficult to solubilize with conventional non-denaturing cell solubilization buffers (RIPA buffer, 1% Triton X-100, etc.). Lipid raft proteins can be extracted with high efficiency from membrane fractions that cannot be solubilized with conventional buffers under mild conditions with low protein denaturation, making it useful for functional analysis of lipid raft proteins.
■What is lipid raft
■Problems with lipid raft analysis
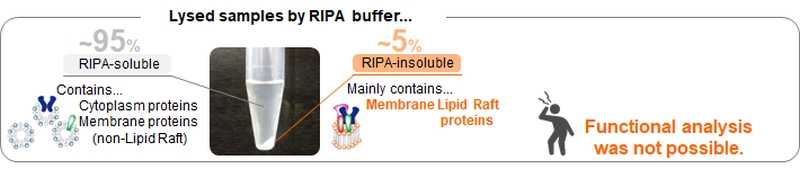
Lipid rafts are difficult to solubilize in detergent buffers (such as 1%Triton X -100) or RIPA buffers, which have a low protein denaturation effect, and are sometimes called detergent resistant membranes (DRM). Sodium dodecyl sulfate (SDS) can solubilize lipid rafts, however the denaturation effect is too strong to do functional analysis.
Features
| Section | SDS buffer | RIPA buffer | UltraRIPA kit |
|---|---|---|---|
| Cytosolic proteins | Extractable but in denatured state | Extractable in native state | Extractable in native state (A-buffer soluble fraction) |
| Membrane proteins (enriched in non-lipid raft) |
Extractable but in denatured state | Extractable in native state | Extractable in native state (A-buffer soluble fraction) |
| Membrane proteins (enriched in lipid raft) |
Extractable but in denatured state | Unable to extract | Extractable in native state (B-buffer soluble fraction) |
| Immunoprecipitation of lipid raft-enriched proteins |
Difficult due to denatured state | Difficult due to unextractable | Applicable because of native state |
| Enzymatic assay of lipid raft-enriched proteins |
Difficult due to denatured state | Difficult due to unextractable | Applicable because of native state |
- Rapid lipid raft protein extraction is possible with simple operation.
- Two buffers (A-buffer, B-buffer) are used for two-step extraction. Cytoplasmic and non-lipid raft enriched proteins are extracted first, followed by lipid raft enriched proteins.
- Both buffers are low in protein denaturation and can be used for functional analysis of proteins.
- A-buffer has the same composition as a standard RIPA buffer*. B-buffer contains surfactant, which can be removed by dialysis.
*1% NP-40 Alternative, 0.1% SDS, 50 mM Tris-HCl (pH8.0), 150 mM NaCl, 0.5% Sodium Deoxycholate - Optimized for mammalian cells and tissues.
- Lysates prepared with this product can be used for enzyme activity assays, immunoprecipitation, protein quantification (BCA assay, dissolved in B-buffer), SDS-PAGE, and western blotting.
Procedure Overview
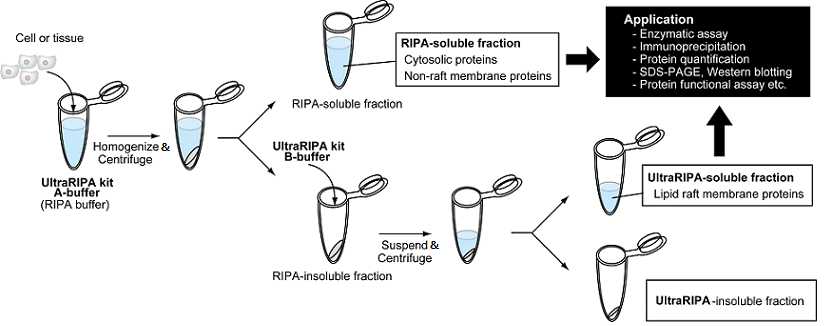
- Dissolve cultured cells or tissues with A-buffer.
*Homogenization or ultrasonication are recommended to increase solubilization efficiency and to prevent nuclear contamination. - Centrifuge samples and separate A-buffer soluble fraction (supernatant) and insoluble fraction (pellet). The supernatant (A-buffer soluble fraction) mainly contains cytosolic proteins and non-lipid raft proteins.
- Add B-buffer to the pellet (A-buffer insoluble fraction) and suspend well.
- Centrifuge and separate into soluble fraction (supernatant) and insoluble fraction (pellet) as in 2. The supernatant (B-buffer soluble fraction) contains lipid raft proteins.
- Use them for further assays.
※ A-buffer and B-buffer cannot be used for the Bradford protein assay. Please use BCA assay to quantify proteins.
※ The product is optimized for two-step extraction using A-buffer and B-buffer, however there are examples where cells were lysed and extracted only with B buffer. For details, see the application data.
Example of Use
Method:
A buffer (RIPA buffer) was added to a whole mouse brain and fractured using a homogenizer and sonicator.
After centrifugation, the precipitates was re-suspended with 2% SDS buffer, RIPA buffer or B buffer (UltraRIPA buffer). The protein content of each supernatant obtained after centrifugation was compared by silver staining and BCA assay.
Results:
B buffer (UltraRIPA buffer) could extracted more than 70% of the RIPA-insoluble proteins.
Kit Contents
|
 |
Application Data
(Data provided by Laboratory of Health Chemistry, Graduate School of Pharmaceutical Sciences, The University of Tokyo)
Composition of each buffer
- SDS buffer: 2% SDS, 1% Triton X-100, 50 mM HEPES・Na (pH 7.2), 150 mM NaCl
- UltraRIPA kit A buffer: 1% NP-40 Alternative, 0.1% SDS, 50 mM Tris-HCl (pH8.0), 150 mM NaCl, 0.5% Sodium Deoxycholate
- UltraRIPA kit B buffer: undisclosed
- 1% Triton X-100: 1% Triton X-100, 50 mM HEPES・Na (pH 7.2), 150 mM NaCl
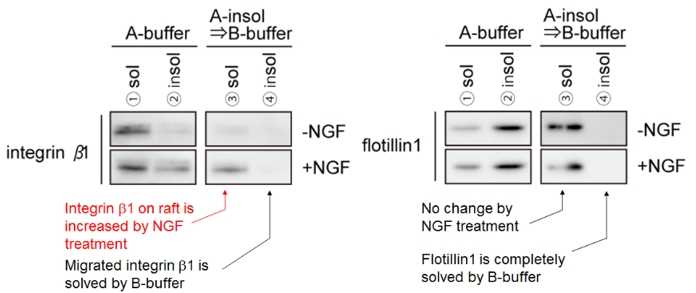
(Data provided by Department of PNS Research National Institute of Neuroscience, NCNP)
Sample: mouse primary cultured DRG neurons (DIV13, ~106 cells/35 mm dish)
Target protein: Integrinβ1, Flotillin 1
Protocol:
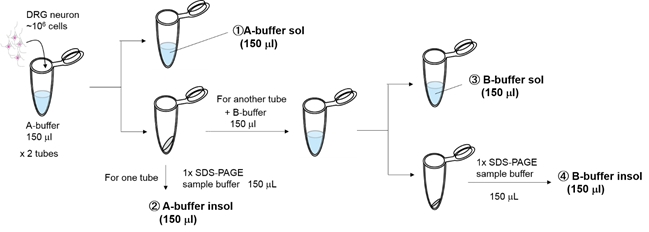
- Culture neurons with or without 50 ng/ml NGF (2 vials each).
- Wash the cells with PBS, add A-buffer(150 μL), and incubate on ice.
- Separate soluble fraction(①A-buffer sol 150ul) and insoluble fraction by centrifugation.
- To one of the two A-buffer insoluble fractions, add 1×SDS sample buffer(150 μL) to completely solubilize(②A-buffer insol 150ul).
- To the other A-buffer insoluble fraction, add B-buffer(150 μL) to suspend the pellet.
- Separate soluble fraction(③B-buffer sol 150 μl) and insoluble fraction by centrifugation.
- To the B-buffer insoluble fraction, add 1×SDS-PAGE sample buffer(150 μL) to completely solubilize(④B-buffer insol 150 μl).
Result:
(Data obtained in collaboration with Professor Akihiko Takashima and Assistant Professor Akio Sumioka, Factory of Science, Gakushuin University)
Verification of solubilization efficiency by direct addition of B-buffer
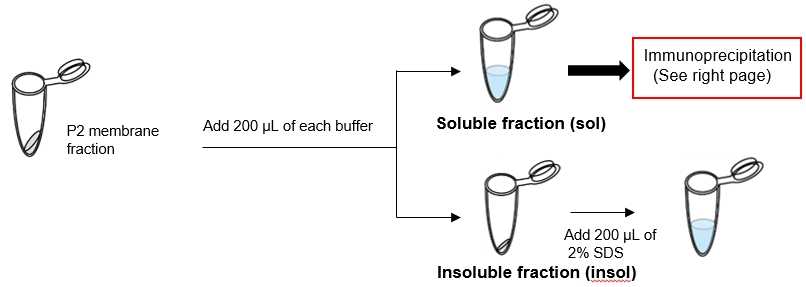
Sample: P2 membrane fraction* derived from mouse brain tissue (hippocampus + cerebral cortex)
*Protocol of extracting P2 membrane fraction
Composition of each buffer
- SDS: 2% SDS, 50 mM Tris-HCl (pH 8.0), 150 mM NaCl
- 1% Triton: 1% TritonX-100, 50 mM Tris-HCl (pH 8.0), 150 mM NaCl
- UltraRIPA kit A buffer (RIPA): 1% NP-40 Alternative, 0.1% SDS, 50 mM Tris-HCl (pH8.0), 150 mM NaCl, 0.5% Sodium Deoxycholate
- UltraRIPA kit B buffer: undisclosed
Protocol:
Result:
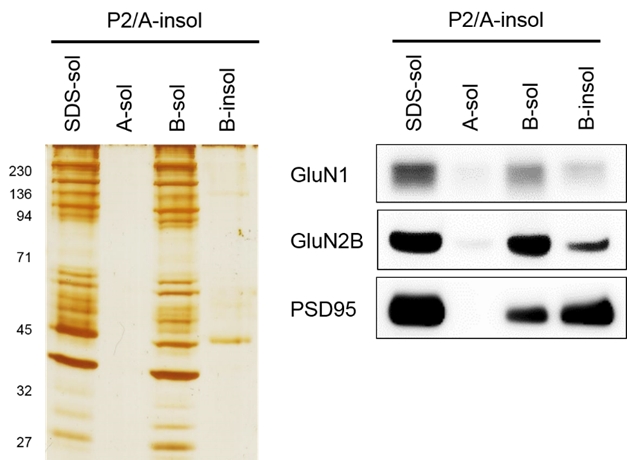
Verification of B-buffer solubilization efficiency of RIPA-Insoluble fractions derived from neuronal plasma membranes
Sample: P2 membrane fraction derived from mouse brain tissue (hippocampus + cerebral cortex)
Protocol:
- Add 200 μl of A-buffer (RIPA) P2 membrane fraction and sonicate on ice.
- Centrifuge the sonicated solution at high speed (×100,000 g) to remove soluble fraction and isolate insoluble fraction.
- To the A-buffer-insoluble fraction, add 200 μl of 2% SDS, A-buffer or B-buffer and sonicate on ice.
- Separated the soluble and insoluble fractions by centrifugation (×100,000 g).
- To determine proteins remaining in the B-buffer-insoluble fraction, add 2% SDS to the B-buffer-insoluble fraction to completely solubilize it.
Result:
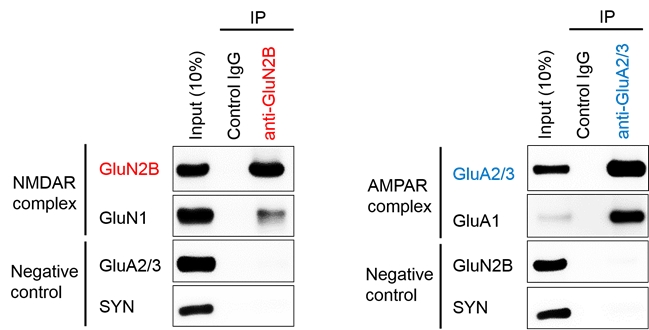
Analysis of neuronal synaptic protein complex
Sample: P2 membrane fraction derived from mouse brain tissue (hippocampus + cerebral cortex)
Target: NMDA glutamate receptor, NMDAR (GluN1/GluN2B)
AMPA glutamate receptor, AMPAR (GluA1/GluA2)
Protocol:
- Add UltraRIPA kit B-buffer to P2 membrane fraction and sonicated on ice.
- Centrifuge the sonocated solution at high speed (×100,000 g) to obtain a soluble fraction.
- To 100 μL of the soluble fraction, add 1 μg of control-IgG or anti-GluN 2B antibody or anti-GluA 2/3 antibody. After 1 hour reaction at 4℃, add 20 μL bed vol of Protein A beads and react for another 1 hour.
- Wash the beads 3 times with PBST (0.05% Tween20 in PBS).
- Add 1× SDS-PAGE sample buffer and elute under heating conditions at 100°C.
Result:
Sample: P2 membrane fraction* derived from mouse brain tissue (hippocampus + cerebral cortex)
*Protocol of extracting P2 membrane fraction
Composition of each buffer
- SDS: 2% SDS, 50 mM Tris-HCl (pH 8.0), 150 mM NaCl
- 1% Triton: 1% TritonX-100, 50 mM Tris-HCl (pH 8.0), 150 mM NaCl
- UltraRIPA kit A buffer (RIPA): 1% NP-40 Alternative, 0.1% SDS, 50 mM Tris-HCl (pH8.0), 150 mM NaCl, 0.5% Sodium Deoxycholate
- UltraRIPA kit B buffer: undisclosed
Protocol:
Result:
Sample: P2 membrane fraction derived from mouse brain tissue (hippocampus + cerebral cortex)
Protocol:
- Add 200 μl of A-buffer (RIPA) P2 membrane fraction and sonicate on ice.
- Centrifuge the sonicated solution at high speed (×100,000 g) to remove soluble fraction and isolate insoluble fraction.
- To the A-buffer-insoluble fraction, add 200 μl of 2% SDS, A-buffer or B-buffer and sonicate on ice.
- Separated the soluble and insoluble fractions by centrifugation (×100,000 g).
- To determine proteins remaining in the B-buffer-insoluble fraction, add 2% SDS to the B-buffer-insoluble fraction to completely solubilize it.
Result:
Analysis of neuronal synaptic protein complex
Sample: P2 membrane fraction derived from mouse brain tissue (hippocampus + cerebral cortex)
Target: NMDA glutamate receptor, NMDAR (GluN1/GluN2B)
AMPA glutamate receptor, AMPAR (GluA1/GluA2)
Protocol:
- Add UltraRIPA kit B-buffer to P2 membrane fraction and sonicated on ice.
- Centrifuge the sonocated solution at high speed (×100,000 g) to obtain a soluble fraction.
- To 100 μL of the soluble fraction, add 1 μg of control-IgG or anti-GluN 2B antibody or anti-GluA 2/3 antibody. After 1 hour reaction at 4℃, add 20 μL bed vol of Protein A beads and react for another 1 hour.
- Wash the beads 3 times with PBST (0.05% Tween20 in PBS).
- Add 1× SDS-PAGE sample buffer and elute under heating conditions at 100°C.
Result:
Sample: P2 membrane fraction derived from mouse brain tissue (hippocampus + cerebral cortex)
Target: NMDA glutamate receptor, NMDAR (GluN1/GluN2B)
AMPA glutamate receptor, AMPAR (GluA1/GluA2)
Protocol:
- Add UltraRIPA kit B-buffer to P2 membrane fraction and sonicated on ice.
- Centrifuge the sonocated solution at high speed (×100,000 g) to obtain a soluble fraction.
- To 100 μL of the soluble fraction, add 1 μg of control-IgG or anti-GluN 2B antibody or anti-GluA 2/3 antibody. After 1 hour reaction at 4℃, add 20 μL bed vol of Protein A beads and react for another 1 hour.
- Wash the beads 3 times with PBST (0.05% Tween20 in PBS).
- Add 1× SDS-PAGE sample buffer and elute under heating conditions at 100°C.
Result:
FAQ
A-1 B-buffer has higher solubilization activity than RIPA (=A-buffer). This kit uses a two-step protocol to collect the RIPA-insoluble fraction and then dissolve it in B-buffer to concentrate and simply purify lipid raft proteins contained in the RIPA-insoluble fraction. When a sample is directly dissolved in B-buffer, the solubilization rate is higher than RIPA.
A-2 This product focuses on the fact that RIPA-insoluble fraction contains a lot of lipid raft proteins. By further solubilizing RIPA-insoluble fraction with B-buffer, it is mainly intended to solubilize lipid raft proteins in a non-denatured state. Therefore, even if proteins are not lipid rafts, if they are insoluble in RIPA buffer, they may be detected by this kit (see Q-3 for nuclear protein contamination).
This kit uses RIPA buffer with high solubilization capacity as A-buffer, however it is possible to replace RIPA buffer with another solubilizing buffer such as 1% TritonX-100.
A-3 This product uses a simple protocol to separate RIPA-soluble and RIPA-insoluble fractions by solubilizing A buffer and then centrifuging. Therefore, if unfractured cells/tissues or nuclei remain, RIPA-insoluble fraction may become contaminated. In particular, RIPA buffer has a weak nuclear solubilization efficiency, so there is a risk that a large amount of nuclei will remain in RIPA-insoluble fraction without physical fragmentation. On the other hand, B-buffer has a high nuclear solubilization efficiency, so if nuclei remain, genomic DNA will be released and the solution will become mushy. When solubilizing with A-buffer, sonication can destroy nuclei and fragment genomic DNA. The use of homogenization or sonication is strongly recommended because nuclear contamination can adversely affect subsequent applications.
A-4 Unfortunately, the composition of B-buffer and surfactant-type are not disclosed. However, our supplier has confirmed from various studies that B-buffer does not affect electrophoresis.
A-5 RIPA buffer can solubilize about 90~95% of total protein, including membrane proteins (depending on tissue and cell type). Therefore, in terms of total protein, the amount of protein contained in RIPA-insoluble fraction is very small, less than 10% of total protein. If RIPA-insoluble fraction cannot be observed visually, it is necessary to increase the number of cells or amount of tissue. If you want to increase the protein concentration in B-buffer soluble fraction, please reduce the volume of B-buffer added to RIPA-insoluble fraction. We recommend the following positive control experiment to consider the conditions.
Positive control experiment for considering starting-amount of sample
Things to prepare: 2% SDS buffer (50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 2% SDS)
Protocol:
- Collect RIPA-insoluble fraction
- Add 2% SDS buffer
- Perform protein assay
- Estimate the total amount of protein contained in the RIPA-insoluble fraction
A-6 We cannot guarantee that proteins detected as lipid raft markers by other methods will be obtained in A-buffer insoluble fraction. In particular, some lipid raft markers may vary depending on cell type, tissue, and the presence of external signals. Please refer to the following for specific examples.
Example 1: Insoluble in 1% TritonX-100, but detected in RIPA-buffer soluble fraction.
Improvement ideas: Because RIPA buffer (1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate) is more extractable than 1% TritonX-100, so target proteins may be solubilized in RIPA buffer. Using 1% TritonX-100 buffer instead of A-buffer(= RIPA buffer) may concentrate and solubilize the target proteins.
Example 2: Insoluble but not precipitated (in the case of insufficient centrifugation)
Improvement ideas: We recommend a centrifugal condition of 10,000×g or more, however it may not be sufficient depending on the target protein. In such cases, centrifuging at 20,000×g may precipitate the proteins. We recommend that you consider the centrifugal conditions.
A-7 Surfactant(s) in B-buffer can be removed by dialysis. The pore size of dialysis membrane should be around 5 kDa. However, some proteins may aggregate or denature when surfactant(s) is removed, so it may be necessary to add another surfactant(s) to dialysis buffer. It is recommended that a preliminary evaluation of dialysis be performed before the assay.
Product Information
[Date : March 08 2026 00:07]
| Detail | Product Name | Product Code | Supplier | Size | Price | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
UltraRIPA kit for Lipid Raft DatasheetThis may not be the latest data sheet. |
F015 | BDLBioDynamics Laboratory Inc | 1 kit | $140 | |||||||||||||||||||||||||||||||
|
|
|
||||||||||||||||||||||||||||||||||
[Date : March 08 2026 00:07]
UltraRIPA kit for Lipid Raft
DatasheetThis may not be the latest data sheet.
- Product Code: F015
- Supplier: BDL
- Size: 1kit
- Price: $140
| Description |
ULTRARIPA kit, can efficiently and rapidly extract membrane proteins or membrane-associated proteins enriched in lipid rafts with native structure and function. |
||
|---|---|---|---|
| Storage | 4°C | CAS | |
| Link |
|
||
Kit Components
[Date : March 08 2026 00:07]
Detail
Product Name
Product Code
Supplier
Size
Price
RIPA Buffer
DatasheetThis may not be the latest data sheet.
F015A-100
BDLBioDynamics Laboratory Inc
100
ml
$100
Description
A buffer widely used for cell lysis and protein extraction.
Storage
4°C
CAS
Link
RIPA Buffer
DatasheetThis may not be the latest data sheet.
F015A-250
BDLBioDynamics Laboratory Inc
250
ml
$200
Description
A buffer widely used for cell lysis and protein extraction.
Storage
4°C
CAS
Link
UltraRIPA Buffer
DatasheetThis may not be the latest data sheet.
F015B
BDLBioDynamics Laboratory Inc
100
ml
$220
Description
Next-generation RIPA buffer that greatly improves the extraction efficiency of membrane proteins. Because proteins can be solubilized under non-denaturing conditions, it can also be used in experiments such as enzyme activity and immunoprecipitation.
Storage
4°C
CAS
Link
[Date : March 08 2026 00:07]
RIPA Buffer
DatasheetThis may not be the latest data sheet.
- Product Code: F015A-100
- Supplier: BDL
- Size: 100ml
-
Price:
$100
Description
A buffer widely used for cell lysis and protein extraction.
Storage
4°C
CAS
Link
RIPA Buffer
DatasheetThis may not be the latest data sheet.
- Product Code: F015A-250
- Supplier: BDL
- Size: 250ml
-
Price:
$200
Description
A buffer widely used for cell lysis and protein extraction.
Storage
4°C
CAS
Link
UltraRIPA Buffer
DatasheetThis may not be the latest data sheet.
- Product Code: F015B
- Supplier: BDL
- Size: 100ml
-
Price:
$220
Description
Next-generation RIPA buffer that greatly improves the extraction efficiency of membrane proteins. Because proteins can be solubilized under non-denaturing conditions, it can also be used in experiments such as enzyme activity and immunoprecipitation.
Storage
4°C
CAS
Link
[Date : March 08 2026 00:07]
| Detail | Product Name | Product Code | Supplier | Size | Price | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
RIPA Buffer DatasheetThis may not be the latest data sheet. |
F015A-100 | BDLBioDynamics Laboratory Inc | 100 ml | $100 | |||||||||||||||||||||||||||||||
|
|
|
||||||||||||||||||||||||||||||||||
|
RIPA Buffer DatasheetThis may not be the latest data sheet. |
F015A-250 | BDLBioDynamics Laboratory Inc | 250 ml | $200 | |||||||||||||||||||||||||||||||
|
|
|
||||||||||||||||||||||||||||||||||
|
UltraRIPA Buffer DatasheetThis may not be the latest data sheet. |
F015B | BDLBioDynamics Laboratory Inc | 100 ml | $220 | |||||||||||||||||||||||||||||||
|
|
|
||||||||||||||||||||||||||||||||||
[Date : March 08 2026 00:07]
RIPA Buffer
DatasheetThis may not be the latest data sheet.
- Product Code: F015A-100
- Supplier: BDL
- Size: 100ml
- Price: $100
| Description |
A buffer widely used for cell lysis and protein extraction. |
||
|---|---|---|---|
| Storage | 4°C | CAS | |
| Link | |||
RIPA Buffer
DatasheetThis may not be the latest data sheet.
- Product Code: F015A-250
- Supplier: BDL
- Size: 250ml
- Price: $200
| Description |
A buffer widely used for cell lysis and protein extraction. |
||
|---|---|---|---|
| Storage | 4°C | CAS | |
| Link | |||
UltraRIPA Buffer
DatasheetThis may not be the latest data sheet.
- Product Code: F015B
- Supplier: BDL
- Size: 100ml
- Price: $220
| Description |
Next-generation RIPA buffer that greatly improves the extraction efficiency of membrane proteins. Because proteins can be solubilized under non-denaturing conditions, it can also be used in experiments such as enzyme activity and immunoprecipitation. |
||
|---|---|---|---|
| Storage | 4°C | CAS | |
| Link | |||
CONTACT
export@funakoshi.co.jp
- ※Prices on our website are for your reference only. Please inquire your distributor for your prices.
- ※Please note that Product Information or Price may change without notice.