10 min! Rapid & Easy DNA Extraction Kaneka Easy DNA Extraction Kit (version 2)
Date:January 26 2017Web Page No:80099

- Features
- Examples of Use
- Protocol Overview
- Various Applications
- Kit Contents
- Citations
- Product Information
Features
- Simple protocol - Just adding reagents to sample, incubate and mix for extracting.
- Time saving - Around 10 minutes per sample for extraction
- Room temperature storage
- This kit can be applied to a wide range of samples from animal tissues and cells to plants. Broad range of sample are compatible with this kit
- For 50 tests. Bulk size for 1,000 tests is also available.
- Sample: animal tissues, cultured cells, microorganisms, plants, blood (heparin or EDTA treatment), feces
- Sample volume: 5-15 μl (blood). For other samples, please refer to protocol overview.
- Cells, Animal Tissue, Plant, Bacteria, Blood and Feces.
Examples of Use
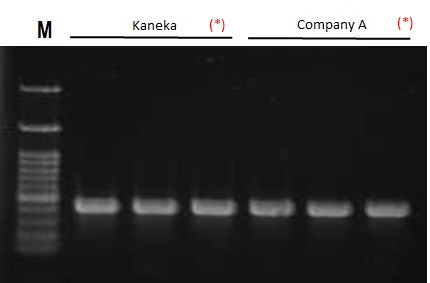
DNAs were extracted from mouse tail tissue samples using extraction kits from each company, and then PCR was performed. Although the operation time was 8 minutes (this product: Kaneka) and 13 minutes (company A), the extraction performance was almost the same.
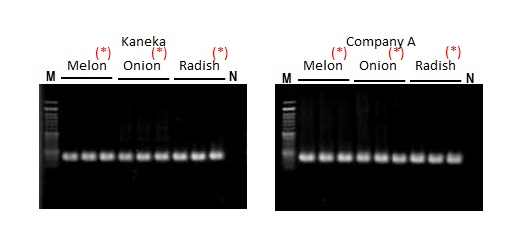
DNAs were extracted from plant seed samples using extraction kits from each company, and then PCR was performed. The operation time was 8 minutes (this product: Kaneka) and 80 minutes (company A). There was no difference in amplification efficiency.
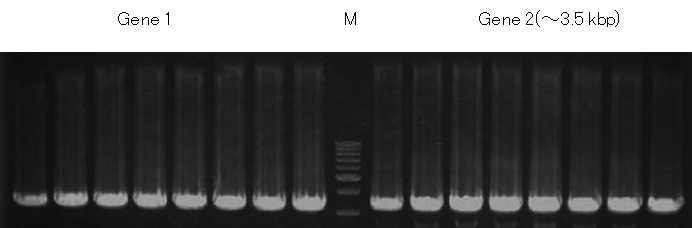
DNAs were extracted using this kit from yeast (Pichia pastoris) colony into which genes were introduced at 2 positions in genome sequence. PCR was then performed to confirm whether gene introduction was successful (target amplification product: approximately 3.5 kb). The extraction process took 8 minutes. The extraction was easily completed without organic solvent treatment.
Protocol Overview

※ The appropriate amount of a sample depends on the type and condition of the specimen.
※ The following reagent amounts are for 50 μl PCR reaction system.
※ Sample-preparation for other samples are also available. Please refer to the data sheet for details.
Animal tissue (mouse tail) sample
- Add a cut mouse tail (5-8 mm) to a PCR tube, add 100 μl of reagent A, and stir well by pipetting.
- Incubate the PCR tube in a heat block at 98°C for 8 minutes.
- After the PCR tube cools, add 14 μl of reagent B, and stir well.
- Stir well the extract obtained in (3) before use, and use 1-5 μl as the template DNA for PCR.
Plant sample
- Add a leaf or seed piece cut into 5-8 mm squares to a PCR tube, add 100 μl of reagent A, and stir well by pipetting.
- Hereinafter, the same as for mouse tail samples.
Blood sample
- Add 5-15 μl of blood with anticoagulant (heparin, EDTA, etc.) to a PCR tube, add 100 μl of reagent A, and stir well by pipetting.
- Incubate the PCR tube in a heat block at 98°C for 8 minutes.
- After the PCR tube cools, add 14 μl of reagent B, and stir well.
- Stir well the extract obtained in A before use, and use 1-5 μl as the template DNA for PCR.
※ If sample-amount is little, please reduce the total volume of the reagent A & B without changing the ratio between reagent A and reagent B.
Various Applications
- Yeast colony (Pichia pastoris)
- Shrimp
- Fish fins
- Cultured cells (HEK293)
- Knockout mouse tissue (tail)
- HIV lentivector cells
- Bird feces
- Candida colony
- Plant seeds
Kit Contents
- Solution A
- Solution B
Citations
- No.: 1
-
Literature information:
Diwan D et al. Ordered genome change of plant and animal body cells revealed by the genome profiling method. FEBS Lett 2016 Jul;590(14):2119-26
Diwan D et al
2016/01/01
-
Remarks:
Sample species: zebrafish, Sample type: microcubes -
URL:
- No.: 2
-
Literature information:
Numamoto M et al. Efficient genome editing by CRISPR/Cas9 with a tRNA-sgRNA fusion in the methylotrophic yeast Ogataea polymorpha. J Biosci Bioeng 2017 Nov;124(5):487-492
Numamoto M et al
2017/01/01
-
Remarks:
Sample species: yeast, Sample type: cell -
URL:
- No.: 3
-
Literature information:
Kitamura M et al. Rapid identification of drug-type strains in Cannabis sativa using loop-mediated isothermal amplification assay. J Nat Med 2017 Jan;71(1):86-95
Kitamura M et al
2017/01/01
-
Remarks:
Sample species: cannabis -
URL:
- No.: 4
-
Literature information:
Yamamuro T et al. Development of rapid and simple method for DNA extraction from cannabis resin based on the evaluation of relative PCR amplification ability. Forensic Sci Int 2018 Jun;287:176-182
Yamamuro T et al
2018/01/01
-
Remarks:
Sample species: cannabis, Sample type: leaves and resin -
URL:
- No.: 5
-
Literature information:
Yamamoto M et al. Rapid Detection of Candida auris Based on Loop-Mediated Isothermal Amplification (LAMP). J Clin Microbiol 2018 Sep;56(9)
Yamamoto M et al
2018/01/01
-
Remarks:
Sample species: human, Sample type: ear swab -
URL:
- No.: 6
-
Literature information:
Kitamura M et al. Rapid screening method for male DNA by using the loop-mediated isothermal amplification assay. Int J Legal Med 2018 Jul;132(4):975-981
Kitamura M et al
2018/01/01
-
Remarks:
Sample species: human, Sample type: blood and saliva stains -
URL:
- No.: 7
-
Literature information:
Chikamatsu K et al. Evaluation of PyroMark Q24 pyrosequencing as a method for the identification of mycobacteria. Diagn Microbiol Infect Dis 2018 Jan;90(1):35-39
Chikamatsu K et al
2018/01/01
-
Remarks:
Sample species: mycobacteria, Sample type: cell -
URL:
- No.: 8
-
Literature information:
Tsukamoto T. Transcriptional gene silencing limits CXCR4-associated depletion of bone marrow CD34+ cells in HIV-1 infection. AIDS 2018 Aug;32(13):1737-1747
Tsukamoto T
2018/01/01
-
Remarks:
Sample species: mouse, Sample type: cell -
URL:
- No.: 9
-
Literature information:
Goto T et al. An Autopsy Case of Rupture of Infectious Thoracic Aortitis Induced by Methicillin-Resistant Staphylococcus Aureus. Am J Case Rep 2019 Nov;20:1612-1618
Goto T et al
2019/01/01
-
Remarks:
Sample species: human, Sample type: blood, tissue -
URL:
- No.: 10
-
Literature information:
Du Y et al. Using CRISPR/Cas9 for Gene Knockout in Immunodeficient NSG Mice. Methods Mol Biol 2019;1874:139-168
Du Y et al
2019/01/01
-
Remarks:
Sample species: mouse -
URL:
- No.: 11
-
Literature information:
Kitamura M et al. Rapid identification of Aconitum plants based on loop-mediated isothermal amplification assay. BMC Res Notes 2019 Jul;12(1):408
Kitamura M et al
2019/01/01
-
Remarks:
Sample species: Aconitum plants, Sample type: leaf -
URL:
- No.: 12
-
Literature information:
Benderli NC et al. Feasibility of microbial sample collection on the skin from people in Yaoundé, Cameroon. Drug Discov Ther 2019;13(6):360-364
Benderli NC et al
2019/01/01
-
Remarks:
Sample species: human, Sample type: skin swab -
URL:
- No.: 13
-
Literature information:
Tsukamoto T. HIV Impacts CD34<sup>+</sup> Progenitors Involved in T-Cell Differentiation During Coculture With Mouse Stromal OP9-DL1 Cells. Front Immunol 2019;10:81
Tsukamoto T
2019/01/01
-
Remarks:
Sample species: human -
URL:
- No.: 14
-
Literature information:
Nomura T et al. Highly Efficient CRISPR-Associated Protein 9 Ribonucleoprotein-Based Genome Editing in <i>Euglena gracilis</i>. STAR Protoc 2020 Jun;1(1):100023
Nomura T et al
2020/01/01
-
Remarks:
Sample species: Euglena gracilis, Sample type: cell -
URL:
- No.: 15
-
Literature information:
Hieno A et al. Detection of the Genus <i>Phytophthora</i> and the Species <i>Phytophthora nicotianae</i> by LAMP with a QProbe. Plant Dis 2020 Sep;104(9):2469-2480
Hieno A et al
2020/01/01
-
Remarks:
Sample species: tobacco NULLTomato and egg-plant, Sample type: stemsNULLfruits -
URL:
- No.: 16
-
Literature information:
Yoshida M et al. A novel DNA chromatography method to discriminate Mycobacterium abscessus subspecies and macrolide susceptibility. EBioMedicine 2021 Feb;64:103187
Yoshida M et al
2021/01/01
-
Remarks:
Sample species: mycobacterial, Sample type: cell -
URL:
- No.: 17
-
Literature information:
Chin KL et al. Identification of a Mycobacterium tuberculosis-specific gene marker for diagnosis of tuberculosis using semi-nested melt-MAMA qPCR (lprM-MAMA). Tuberculosis (Edinb) 2020 Dec;125:102003
Chin KL et al
2020/01/01
-
Remarks:
Sample species: human, Sample type: sputum -
URL:
- No.: 18
-
Literature information:
But GW et al. Rapid detection of CITES-listed shark fin species by loop-mediated isothermal amplification assay with potential for field use. Sci Rep 2020 Mar;10(1):4455
But GW et al
2020/01/01
-
Remarks:
Sample species: shark, fish, Sample type: dried processed shark fin samples, frozen tissues, inner organs -
URL:
- No.: 19
-
Literature information:
Boytard L et al. Lung-derived HMGB1 is detrimental for vascular remodeling of metabolically imbalanced arterial macrophages. Nat Commun 2020 08;11(1):4311
Boytard L et al
2020/01/01
-
Remarks:
Sample species: mouse, Sample type: BMDM(骨髄由来マクロファージ) -
URL:
- No.: 20
-
Literature information:
Horibata Y et al. Locations and contributions of the phosphotransferases EPT1 and CEPT1 to the biosynthesis of ethanolamine phospholipids. J Lipid Res 2020 Aug;61(8):1221-1231
Horibata Y et al
2020/01/01
-
Remarks:
Sample species: human, Sample type: HEK293 cells -
URL:
- No.: 21
-
Literature information:
Hagihara Y et al. Primordial germ cell-specific expression of eGFP in transgenic chickens. Genesis 2020 Aug;58(8):e23388
Hagihara Y et al
2020/01/01
-
Remarks:
Sample species: chickens, Sample type: cell -
URL:
- No.: 22
-
Literature information:
Nagai K et al. Antagonistic regulation of the gibberellic acid response during stem growth in rice. Nature 2020 Aug;584(7819):109-114
Nagai K et al
2020/01/01
-
Remarks:
-
URL:
- No.: 23
-
Literature information:
Doi H et al. On-site environmental DNA detection of species using ultrarapid mobile PCR. Mol Ecol Resour 2021 Oct;21(7):2364-2368
Doi H et al
2021/01/01
-
Remarks:
Sample species: fish, Sample type: water -
URL:
- No.: 24
-
Literature information:
Soejima M et al. High-resolution melting analysis for detection of fusion allele of FUT2. Electrophoresis 2021 Feb;42(3):315-318
Soejima M et al
2021/01/01
-
Remarks:
-
URL:
- No.: 25
-
Literature information:
Fukuzawa T et al. Filtration extraction method using a microfluidic channel for measuring environmental DNA. Mol Ecol Resour 2022 Oct;22(7):2651-2661
Fukuzawa T et al
2022/01/01
-
Remarks:
Sample species: eDNA, Sample type: water -
URL:
- No.: 26
-
Literature information:
Igarashi Y et al. Development of a nucleic acid chromatography assay for the detection of commonly isolated rapidly growing mycobacteria. J Med Microbiol 2021 Dec;70(12)
Igarashi Y et al
2021/01/01
-
Remarks:
Sample species: mycobacterial, Sample type: cell -
URL:
- No.: 27
-
Literature information:
Fujiyoshi S et al. Suitcase Lab: new, portable, and deployable equipment for rapid detection of specific harmful algae in Chilean coastal waters. Environ Sci Pollut Res Int 2021 Mar;28(11):14144-14155
Fujiyoshi S et al
2021/01/01
-
Remarks:
Sample species: A. pacificum, A. catenella, Sample type: cell -
URL:
- No.: 28
-
Literature information:
Yamamuro T et al. Rapid identification of drug-type and fiber-type cannabis by allele specific duplex PCR. Forensic Sci Int 2021 Jan;318:110634
Yamamuro T et al
2021/01/01
-
Remarks:
Sample species: cannabis, Sample type: seed -
URL:
- No.: 29
-
Literature information:
Horibata Y et al. Differential contributions of choline phosphotransferases CPT1 and CEPT1 to the biosynthesis of choline phospholipids. J Lipid Res 2021;62:100100
Horibata Y et al
2021/01/01
-
Remarks:
Sample species: human, Sample type: HEK293 cells -
URL:
- No.: 30
-
Literature information:
Kawato Y et al. Isolation and characterization of hirame aquareovirus (HAqRV): A new Aquareovirus isolated from diseased hirame Paralichthys olivaceus. Virology 2021 Jul;559:120-130
Kawato Y et al
2021/01/01
-
Remarks:
Sample species: Japanese flounder, Sample type: cell(HINAE cells) -
URL:
- No.: 31
-
Literature information:
Nakanishi H et al. A method for determining the origin of crude drugs derived from animals using MinION, a compact next-generation sequencer. Int J Legal Med 2023 Mar;137(2):581-586
Nakanishi H et al
2023/01/01
-
Remarks:
Sample species: Rangifer tarandus, Sample type: antler velvet -
URL:
- No.: 32
-
Literature information:
Takagi H et al. Influence of mutation in the regulatory domain of α-isopropylmalate synthase from Saccharomyces cerevisiae on its activity and feedback inhibition. Biosci Biotechnol Biochem 2022 May;86(6):755-762
Takagi H et al
2022/01/01
-
Remarks:
-
URL:
- No.: 33
-
Literature information:
Misaki R et al. Establishment of serum-free adapted Chinese hamster ovary cells with double knockout of GDP-mannose-4,6-dehydratase and GDP-fucose transporter. Cytotechnology 2022 Feb;74(1):163-179
Misaki R et al
2022/01/01
-
Remarks:
Sample species: Chinese hamster, Sample type: cell -
URL:
- No.: 34
-
Literature information:
Li A et al. Cytosine base editing systems with minimized off-target effect and molecular size. Nat Commun 2022 Aug;13(1):4531
Li A et al
2022/01/01
-
Remarks:
Sample species: human, Sample type: HEK293T cells -
URL:
- No.: 35
-
Literature information:
Pan X et al. Piperlongumine increases the sensitivity of bladder cancer to cisplatin by mitochondrial ROS. J Clin Lab Anal 2022 Jun;36(6):e24452
Pan X et al
2022/01/01
-
Remarks:
Sample species: human, Sample type: bladder cancer cell -
URL:
- No.: 36
-
Literature information:
Ogai K et al. Skin microbiome profile of healthy Cameroonians and Japanese. Sci Rep 2022 Jan;12(1):1364
Ogai K et al
2022/01/01
-
Remarks:
Sample species: humam, Sample type: Skin swabs -
URL:
- No.: 37
-
Literature information:
Hamamoto H et al. Establishment of a polymerase chain reaction-based method for strain-level management of <i>Enterococcus faecalis</i> EF-2001 using species-specific sequences identified by whole genome sequences. Front Microbiol 2022;13:959063
Hamamoto H et al
2022/01/01
-
Remarks:
Sample species: Lactic acid bacteria, Sample type: cell -
URL:
- No.: 38
-
Literature information:
Nishi T et al. One-Step <i>In Vivo</i> Assembly of Multiple DNA Fragments and Genomic Integration in <i>Komagataella phaffii</i>. ACS Synth Biol 2022 Feb;11(2):644-654
Nishi T et al
2022/01/01
-
Remarks:
Sample species: yeast, Sample type: cell -
URL:
- No.: 39
-
Literature information:
Hisano H et al. Regulation of germination by targeted mutagenesis of grain dormancy genes in barley. Plant Biotechnol J 2022 Jan;20(1):37-46
Hisano H et al
2022/01/01
-
Remarks:
Sample species: barley, Sample type: immature embryos -
URL:
- No.: 40
-
Literature information:
Suga H et al. A Novel Medium for Isolating Two Japanese Species in the <i>Fusarium graminearum</i> Species Complex and a Dipstick DNA Chromatography Assay for Species Identification and Trichothecene Typing. J Fungi (Basel) 2022 Oct;8(10)
Suga H et al
2022/01/01
-
Remarks:
Sample species: Fungal, Sample type: aerial mycelia -
URL:
- No.: 41
-
Literature information:
Imanishi I et al. Antibiotic-resistant status and pathogenic clonal complex of canine Streptococcus canis-associated deep pyoderma. BMC Vet Res 2022 Nov;18(1):395
Imanishi I et al
2022/01/01
-
Remarks:
Sample species: S. canis, Sample type: cell -
URL:
- No.: 42
-
Literature information:
Suzuki T et al. Mutant KRAS drives metabolic reprogramming and autophagic flux in premalignant pancreatic cells. Cancer Gene Ther 2022 May;29(5):505-518
Suzuki T et al
2022/01/01
-
Remarks:
Sample species: human, Sample type: cell(HPNE cells) -
URL:
- No.: 43
-
Literature information:
Narisawa T et al. Fibroblast growth factor receptor type 4 as a potential therapeutic target in clear cell renal cell carcinoma. BMC Cancer 2023 Feb;23(1):170
Narisawa T et al
2023/01/01
-
Remarks:
Sample species: human, Sample type: RCC cells -
URL:
- No.: 44
-
Literature information:
Azuma N et al. Integrator complex subunit 15 controls mRNA splicing and is critical for eye development. Hum Mol Genet 2023 Jun;32(12):2032-2045
Azuma N et al
2023/01/01
-
Remarks:
Sample species: mouse, Sample type: fingers -
URL:
- No.: 45
-
Literature information:
Watanabe K et al. Myeloid-associated differentiation marker is an essential host factor for human parechovirus PeV-A3 entry. Nat Commun 2023 Mar;14(1):1817
Watanabe K et al
2023/01/01
-
Remarks:
Sample species: human, Sample type: cell(HuTu-80) -
URL:
- No.: 46
-
Literature information:
Tamaki S et al. Zeaxanthin is required for eyespot formation and phototaxis in Euglena gracilis. Plant Physiol 2023 Apr;191(4):2414-2426
Tamaki S et al
2023/01/01
-
Remarks:
Sample species: E. gracilis, Sample type: cell -
URL:
- No.: 47
-
Literature information:
CXCR4-associated depletion of bone marrow CD34+ cells following CCR5-tropic HIV-1 infection of humanized NOD/SCID/JAK3null mice and partial protection of those cells by a promotor-targeting shRNA
-
Remarks:
Sample species: mouse, Sample type: cell -
URL:
- No.: 48
-
Literature information:
HIV disrupts CD34+ 1 progenitors involved in T-cell differentiation
-
Remarks:
Sample species: human, Sample type: cell(CD34+ cells) -
URL:
- No.: 49
-
Literature information:
Does simultaneous and sympatric reproduction between two native spined loaches lead to reproductive interference and local extinction?
-
Remarks:
Sample species: fish(spined loaches), Sample type: juvenile specimen -
URL:
- No.: 50
-
Literature information:
Rapid detection of Phytophthora nicotianae by simple DNA extraction and real-time loop-mediated isothermal amplification assay
-
Remarks:
Sample species: plants -
URL:
- No.: 51
-
Literature information:
A simple loop-mediated isothermal amplification assay to detect Phytophthoracolocasiae in infected taro plants
-
Remarks:
Sample species: plants, Sample type: leaf (taro) -
URL:
- No.: 52
-
Literature information:
On-site eDNA detection of species using ultra-rapid mobile PCR
-
Remarks:
Sample species: carp, Sample type: weter -
URL:
- No.: 53
-
Literature information:
Simple and on-site DNA purification for LAMP reaction applicable to non-adult tephritid fruit fly (Diptera: Tephiritidae)
-
Remarks:
Sample species: fruit flies, Sample type: body fluids,eggs -
URL:
- No.: 54
-
Literature information:
Identification of a Rothia mucilaginosa Strain in a Clinical Specimen Based on PCR-Sequencing with Mycobacterium hsp65 Primers.
-
Remarks:
Sample species: human, Sample type: sputum -
URL:
- No.: 55
-
Literature information:
Dominant honeybee colony infestation by Varroa destructor (Acari: Varroidae) K haplotype in Japan
-
Remarks:
Sample species: mites, Sample type: -
URL:
- No.: 56
-
Literature information:
Evaluation of a novel and rapid screening method for the detection of contaminated colistin-resistant bacteria in food
-
Remarks:
Sample species: pork and chicken, Sample type: -
URL:
- No.: 57
-
Literature information:
Rapid identification of Colchicum autumnale based on loop-mediated isothermal amplification (LAMP) assay
-
Remarks:
Sample species: human -
URL:
- No.: 58
-
Literature information:
Rapid and Practical Screening Method for the Detection of Colistin-Resistant Bacteria in Food
-
Remarks:
Sample species: pork and chicken, Sample type: -
URL:
- No.: 59
-
Literature information:
DNA testing of suspected cannabis samples with exceptional morphology using a simple detection kit
-
Remarks:
Sample species: cannabis, Sample type: leaves -
URL:
- No.: 60
-
Literature information:
Molecular analysis of voltage-gated sodium channels to assess τ-fluvalinate resistance in Japanese populations of Varroa destructor (Acari: Varroidae)
-
Remarks:
-
URL:
- No.: 61
-
Literature information:
Toxicity testing of 2,3,7,8-tetrachlorodibenzo-p-dioxin in early-life stage of Japanese medaka: Optimization of conditions for assessing relative potencies of dioxin-like compounds
-
Remarks:
Sample species: fish, Sample type: tail (medaka) -
URL:
- No.: 62
-
Literature information:
徳島県那賀川水系から得られたイワナとアマゴの交雑個体
-
Remarks:
Sample species: fish, Sample type: fin -
URL:
- No.: 63
-
Literature information:
Environmental DNA extraction method for a high and stable DNA yield
-
Remarks:
Sample species: eDNA, Sample type: water -
URL:
- No.: 64
-
Literature information:
The chemical and microbiological characteristics of Awa-bancha, Japanese post-fermented tea
-
Remarks:
-
URL:
- No.: 65
-
Literature information:
Development of Simple Detection Methods of Plant Pathogenic Oomycetes
-
Remarks:
Sample species: planttaro, Sample type: leaves -
URL:
- No.: 66
-
Literature information:
First Report of Pythium myriotylum Causing Stem and Root Rot of Kidney Bean in Taiwan
-
Remarks:
Sample species: Kidney beans, Sample type: stem and root -
URL:
Product Information
[Date : February 23 2026 00:07]
| Detail | Product Name | Product Code | Supplier | Size | Price | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Kaneka Easy DNA Extraction Kit Version2 DatasheetThis may not be the latest data sheet. |
KN-T110005 | KNK | 1 kit | $74 | |||||||||||||||||||||||||||||||
|
|
|
||||||||||||||||||||||||||||||||||
|
Kaneka Easy DNA Extraction Kit version 2 (Bulk for 1,000 tests) |
KN-B101005 | KNK | 1 kit | $1,040 | |||||||||||||||||||||||||||||||
|
|
|
||||||||||||||||||||||||||||||||||
[Date : February 23 2026 00:07]
Kaneka Easy DNA Extraction Kit Version2
DatasheetThis may not be the latest data sheet.
- Product Code: KN-T110005
- Supplier: KNK
- Size: 1kit
- Price: $74
| Description |
This kit can extract template DNA usable for PCR and real-time PCR from biological samples with a simpler operation than conventional methods. |
||
|---|---|---|---|
| Storage | RT | CAS | |
| Link |
|
||
Kaneka Easy DNA Extraction Kit version 2 (Bulk for 1,000 tests)
- Product Code: KN-B101005
- Supplier: KNK
- Size: 1kit
- Price: $1,040
| Description |
This kit can extract template DNA usable for PCR and real-time PCR from biological samples with a simpler operation than conventional methods. |
||
|---|---|---|---|
| Storage | RT | CAS | |
| Link |
|
||
You may also like

CONTACT
export@funakoshi.co.jp
- ※Prices on our website are for your reference only. Please inquire your distributor for your prices.
- ※Please note that Product Information or Price may change without notice.