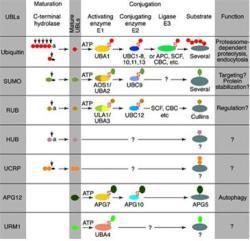

GTX24752.Figure2.jpgMost modifiers mature by proteolytic processing from inactive precursors (a; amino acid). Arrowheads point to the cleavage sites. Ubiquitin is expressed either as polyubiquitin or as a fusion with ribosomal proteins. Conjugation requires activating (E1) and conjugating (E2) enzymes that form thiolesters (S) with the modifiers. Modification of cullins by RUB involves SCF(SKP1/cullin-1/F-box protein) /CBC(cullin-2/elongin B/elonginC) -like E3 enzymes that are also involved in ubiquitination. In contrast to ubiquitin, the UBLs do not seem to form multi-UBL chains. UCRP(ISG15) resembles two ubiquitin moieties linked head-to-tail. Whether HUB1 functions as a modifier is currently unclear. APG12 and URM1 are distinct from the other modifiers because they are unrelated in sequence to ubiquitin. Data contributed by S.Jentsch.
GTX24752 WB Image
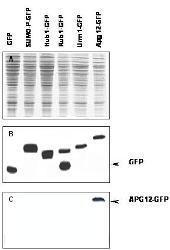
Western blot of APG12 fusion protein. Anti-APG12 antibody generated by immunization with recombinant yeast APG12 was tested by western blot against yeast lysates expressing the APG12-GFP fusion protein and other UBL fusion proteins. All UBLs possess limited homology to Ubiquitin and to each other, therefore it is important to know the degree of reactivity of each antibody against each UBL. Panel A shows total protein staining using ponceau. Panel B shows positions of free GFP or GFP containing recombinant proteins present in each lysate preparation after reaction with a 1:1,000 dilution of GeneTex's anti-GFP